Structural and functional evolution of positively selected sites in pine glutathione S-transferase enzyme family
- PMID: 23846689
- PMCID: PMC3750144
- DOI: 10.1074/jbc.M113.456863
Structural and functional evolution of positively selected sites in pine glutathione S-transferase enzyme family
Abstract
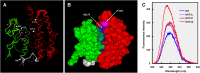
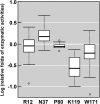
Phylogenetic analyses have identified positive selection as an important driver of protein evolution, both structural and functional. However, the lack of appropriate combined functional and structural assays has generally hindered attempts to elucidate patterns of positively selected sites and their effects on enzyme activity and substrate specificity. In this study we investigated the evolutionary divergence of the glutathione S-transferase (GST) family in Pinus tabuliformis, a pine that is widely distributed from northern to central China, including cold temperate and drought-stressed regions. GSTs play important roles in plant stress tolerance and detoxification. We cloned 44 GST genes from P. tabuliformis and found that 26 of the 44 belong to the largest (Tau) class of GSTs and are differentially expressed across tissues and developmental stages. Substitution models identified five positively selected sites in the Tau GSTs. To examine the functional significance of these positively selected sites, we applied protein structural modeling and site-directed mutagenesis. We found that four of the five positively selected sites significantly affect the enzyme activity and specificity; thus their variation broadens the GST family substrate spectrum. In addition, positive selection has mainly acted on secondary substrate binding sites or sites close to (but not directly at) the primary substrate binding site; thus their variation enables the acquisition of new catalytic functions without compromising the protein primary biochemical properties. Our study sheds light on selective aspects of the functional and structural divergence of the GST family in pine and other organisms.
Keywords: Enzyme Mutation; Molecular Evolution; Plant Biochemistry; Protein Evolution; Protein Structure.
Figures



Similar articles
-
Catalytic properties of glutathione-binding residues in a tau class glutathione transferase (PtGSTU1) from Pinus tabulaeformis.FEBS Lett. 2005 May 9;579(12):2657-62. doi: 10.1016/j.febslet.2005.03.086. Epub 2005 Apr 9. FEBS Lett. 2005. PMID: 15862305
-
Glutathione S-transferase (GST) family in barley: identification of members, enzyme activity, and gene expression pattern.J Plant Physiol. 2013 Sep 15;170(14):1277-84. doi: 10.1016/j.jplph.2013.04.005. Epub 2013 May 10. J Plant Physiol. 2013. PMID: 23664583
-
Functional divergence of the glutathione S-transferase supergene family in Physcomitrella patens reveals complex patterns of large gene family evolution in land plants.Plant Physiol. 2013 Feb;161(2):773-86. doi: 10.1104/pp.112.205815. Epub 2012 Nov 27. Plant Physiol. 2013. PMID: 23188805 Free PMC article.
-
Structure, function and evolution of glutathione transferases: implications for classification of non-mammalian members of an ancient enzyme superfamily.Biochem J. 2001 Nov 15;360(Pt 1):1-16. doi: 10.1042/0264-6021:3600001. Biochem J. 2001. PMID: 11695986 Free PMC article. Review.
-
Glutathione S-transferases--a review.Curr Med Chem. 1999 Apr;6(4):279-309. Curr Med Chem. 1999. PMID: 10101214 Review.
Cited by
-
Genome-Wide Identification, Classification, and Expression Divergence of Glutathione-Transferase Family in Brassica rapa under Multiple Hormone Treatments.Biomed Res Int. 2018 May 24;2018:6023457. doi: 10.1155/2018/6023457. eCollection 2018. Biomed Res Int. 2018. PMID: 29992155 Free PMC article.
-
Adaptive molecular evolution of the two-pore channel 1 gene TPC1 in the karst-adapted genus Primulina (Gesneriaceae).Ann Bot. 2016 Dec;118(7):1257-1268. doi: 10.1093/aob/mcw168. Epub 2016 Aug 30. Ann Bot. 2016. PMID: 27582362 Free PMC article.
-
Molecular cloning, characterization and positively selected sites of the glutathione S-transferase family from Locusta migratoria.PLoS One. 2014 Dec 8;9(12):e114776. doi: 10.1371/journal.pone.0114776. eCollection 2014. PLoS One. 2014. PMID: 25486043 Free PMC article.
-
Interspecific Plastome Recombination Reflects Ancient Reticulate Evolution in Picea (Pinaceae).Mol Biol Evol. 2017 Jul 1;34(7):1689-1701. doi: 10.1093/molbev/msx111. Mol Biol Evol. 2017. PMID: 28383641 Free PMC article.
-
Integrated structural and evolutionary analysis reveals common mechanisms underlying adaptive evolution in mammals.Proc Natl Acad Sci U S A. 2020 Mar 17;117(11):5977-5986. doi: 10.1073/pnas.1916786117. Epub 2020 Mar 2. Proc Natl Acad Sci U S A. 2020. PMID: 32123117 Free PMC article.
References
-
- Barkman T. J., Martins T. R., Sutton E., Stout J. T. (2007) Positive selection for single amino acid change promotes substrate discrimination of a plant volatile-producing enzyme. Mol. Biol. Evol. 24, 1320–1329 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

