Inhibition of protein kinase C signaling maintains rat embryonic stem cell pluripotency
- PMID: 23846691
- PMCID: PMC3750137
- DOI: 10.1074/jbc.M113.455725
Inhibition of protein kinase C signaling maintains rat embryonic stem cell pluripotency
Abstract
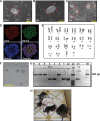
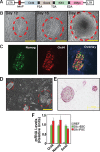
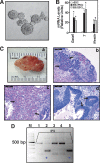
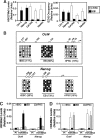
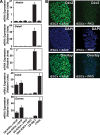
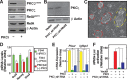
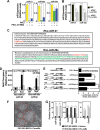
Embryonic stem cell (ESC) pluripotency is orchestrated by distinct signaling pathways that are often targeted to maintain ESC self-renewal or their differentiation to other lineages. We showed earlier that inhibition of PKC signaling maintains pluripotency in mouse ESCs. Therefore, in this study, we investigated the importance of protein kinase C signaling in the context of rat ESC (rESC) pluripotency. Here we show that inhibition of PKC signaling is an efficient strategy to establish and maintain pluripotent rESCs and to facilitate reprogramming of rat embryonic fibroblasts to rat induced pluripotent stem cells. The complete developmental potential of rESCs was confirmed with viable chimeras and germ line transmission. Our molecular analyses indicated that inhibition of a PKCζ-NF-κB-microRNA-21/microRNA-29 regulatory axis contributes to the maintenance of rESC self-renewal. In addition, PKC inhibition maintains ESC-specific epigenetic modifications at the chromatin domains of pluripotency genes and, thereby, maintains their expression. Our results indicate a conserved function of PKC signaling in balancing self-renewal versus differentiation of both mouse and rat ESCs and indicate that targeting PKC signaling might be an efficient strategy to establish ESCs from other mammalian species.
Keywords: Chromatin Modification; Embryonic Stem Cell; NF-κB; PKC; Rat.
Figures

Similar articles
-
Self-renewal versus lineage commitment of embryonic stem cells: protein kinase C signaling shifts the balance.Stem Cells. 2011 Apr;29(4):618-28. doi: 10.1002/stem.605. Stem Cells. 2011. PMID: 21308862 Free PMC article.
-
Ethyl-p-methoxycinnamate enhances oct4 expression and reinforces pluripotency through the NF-κB signaling pathway.Biochem Pharmacol. 2020 Jul;177:113984. doi: 10.1016/j.bcp.2020.113984. Epub 2020 Apr 18. Biochem Pharmacol. 2020. PMID: 32311348
-
Protein kinase C regulates human pluripotent stem cell self-renewal.PLoS One. 2013;8(1):e54122. doi: 10.1371/journal.pone.0054122. Epub 2013 Jan 21. PLoS One. 2013. PMID: 23349801 Free PMC article.
-
[The effect of microRNAs on the regulatory network of pluripotency in embryonic stem cells].Yi Chuan. 2013 Oct;35(10):1153-66. doi: 10.3724/sp.j.1005.2013.01153. Yi Chuan. 2013. PMID: 24459889 Review. Chinese.
-
The Key Role of MicroRNAs in Self-Renewal and Differentiation of Embryonic Stem Cells.Int J Mol Sci. 2020 Aug 31;21(17):6285. doi: 10.3390/ijms21176285. Int J Mol Sci. 2020. PMID: 32877989 Free PMC article. Review.
Cited by
-
MicroRNA‑29a promotes the neural differentiation of rat neural stem/progenitor cells by targeting KLF4.Mol Med Rep. 2020 Aug;22(2):1008-1016. doi: 10.3892/mmr.2020.11177. Epub 2020 May 22. Mol Med Rep. 2020. PMID: 32468029 Free PMC article.
-
Protein kinases and associated pathways in pluripotent state and lineage differentiation.Curr Stem Cell Res Ther. 2014;9(5):366-87. doi: 10.2174/1574888x09666140616130217. Curr Stem Cell Res Ther. 2014. PMID: 24998240 Free PMC article. Review.
-
Urine-derived stem cells in neurological diseases: current state-of-the-art and future directions.Front Mol Neurosci. 2023 Oct 30;16:1229728. doi: 10.3389/fnmol.2023.1229728. eCollection 2023. Front Mol Neurosci. 2023. PMID: 37965041 Free PMC article. Review.
-
Nucleosome remodeling and deacetylation complex and MBD3 influence mouse embryonic stem cell naïve pluripotency under inhibition of protein kinase C.Cell Death Discov. 2022 Aug 1;8(1):344. doi: 10.1038/s41420-022-01131-0. Cell Death Discov. 2022. PMID: 35915078 Free PMC article.
-
Dynamic stem cell states: naive to primed pluripotency in rodents and humans.Nat Rev Mol Cell Biol. 2016 Mar;17(3):155-69. doi: 10.1038/nrm.2015.28. Epub 2016 Feb 10. Nat Rev Mol Cell Biol. 2016. PMID: 26860365 Review.
References
-
- Jacob H. J., Kwitek A. E. (2002) Rat genetics. Attaching physiology and pharmacology to the genome. Nat. Rev. Genet. 3, 33–42 - PubMed
-
- Buehr M., Meek S., Blair K., Yang J., Ure J., Silva J., McLay R., Hall J., Ying Q. L., Smith A. (2008) Capture of authentic embryonic stem cells from rat blastocysts. Cell 135, 1287–1298 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

