DEAF1 is a Pellino1-interacting protein required for interferon production by Sendai virus and double-stranded RNA
- PMID: 23846693
- PMCID: PMC3750155
- DOI: 10.1074/jbc.M113.479550
DEAF1 is a Pellino1-interacting protein required for interferon production by Sendai virus and double-stranded RNA
Abstract
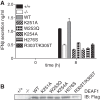
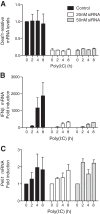
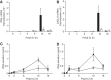
Double-stranded (ds) RNA of viral origin, a ligand for Melanoma Differentiation-associated gene 5 (MDA5) and Toll-Like Receptor 3 (TLR3), induces the TANK-Binding Kinase 1 (TBK1)-dependent phosphorylation and activation of Interferon Regulatory Factor 3 (IRF3) and the E3 ubiquitin ligase Pellino1, which are required for interferon β (IFNβ) gene transcription. Here, we report that Pellino1 interacts with the transcription factor Deformed Epidermal Autoregulatory Factor 1 (DEAF1). The interaction is independent of the E3 ligase activity of Pellino1, but weakened by the phosphorylation of Pellino1. We show that DEAF1 binds to the IFNβ promoter and to IRF3 and IRF7, that it is required for the transcription of the IFNβ gene and IFNβ secretion in MEFs infected with Sendai virus or transfected with poly(I:C). DEAF1 is also needed for TLR3-dependent IFNβ production. Taken together, our results identify DEAF1 as a novel component of the signal transduction network by which dsRNA of viral origin stimulates IFNβ production.
Keywords: Cytokines/Interferon; DEAF1; Double-stranded RNA Viruses; Fibroblast; IRF3; Interferon; Pellino1; TBK1; Transcription Factors.
Figures





Similar articles
-
Pellino1 is required for interferon production by viral double-stranded RNA.J Biol Chem. 2012 Oct 5;287(41):34825-35. doi: 10.1074/jbc.M112.367557. Epub 2012 Aug 17. J Biol Chem. 2012. PMID: 22902624 Free PMC article.
-
Identification of TBK1 complexes required for the phosphorylation of IRF3 and the production of interferon β.Biochem J. 2017 Mar 15;474(7):1163-1174. doi: 10.1042/BCJ20160992. Biochem J. 2017. PMID: 28159912 Free PMC article.
-
MAVS ubiquitination by the E3 ligase TRIM25 and degradation by the proteasome is involved in type I interferon production after activation of the antiviral RIG-I-like receptors.BMC Biol. 2012 May 24;10:44. doi: 10.1186/1741-7007-10-44. BMC Biol. 2012. PMID: 22626058 Free PMC article.
-
Transfected poly(I:C) activates different dsRNA receptors, leading to apoptosis or immunoadjuvant response in androgen-independent prostate cancer cells.J Biol Chem. 2015 Feb 27;290(9):5470-83. doi: 10.1074/jbc.M114.601625. Epub 2015 Jan 7. J Biol Chem. 2015. PMID: 25568326 Free PMC article.
-
Induction and function of IFNβ during viral and bacterial infection.Crit Rev Immunol. 2011;31(6):459-74. doi: 10.1615/critrevimmunol.v31.i6.20. Crit Rev Immunol. 2011. PMID: 22321107 Free PMC article. Review.
Cited by
-
The Emerging Roles of Pellino Family in Pattern Recognition Receptor Signaling.Front Immunol. 2022 Feb 7;13:728794. doi: 10.3389/fimmu.2022.728794. eCollection 2022. Front Immunol. 2022. PMID: 35197966 Free PMC article. Review.
-
The roles of Pellino E3 ubiquitin ligases in immunity.Nat Rev Immunol. 2014 Feb;14(2):122-31. doi: 10.1038/nri3599. Epub 2014 Jan 21. Nat Rev Immunol. 2014. PMID: 24445667 Review.
-
Regulatory role of microRNAs in virus-mediated inflammation.J Inflamm (Lond). 2024 Nov 4;21(1):43. doi: 10.1186/s12950-024-00417-7. J Inflamm (Lond). 2024. PMID: 39497125 Free PMC article. Review.
-
Zinc finger myeloid Nervy DEAF-1 type (ZMYND) domain containing proteins exert molecular interactions to implicate in carcinogenesis.Discov Oncol. 2022 Dec 15;13(1):139. doi: 10.1007/s12672-022-00597-9. Discov Oncol. 2022. PMID: 36520265 Free PMC article. Review.
-
E3 ubiquitin ligases Pellinos as regulators of pattern recognition receptor signaling and immune responses.Immunol Rev. 2015 Jul;266(1):109-22. doi: 10.1111/imr.12298. Immunol Rev. 2015. PMID: 26085210 Free PMC article. Review.
References
-
- Grosshans J., Schnorrer F., Nüsslein-Volhard C. (1999) Oligomerisation of Tube and Pelle leads to nuclear localisation of dorsal. Mech. Dev. 81, 127–138 - PubMed
-
- Haghayeghi A., Sarac A., Czerniecki S., Grosshans J., Schöck F. (2010) Pellino enhances innate immunity in Drosophila. Mech. Dev. 127, 301–307 - PubMed
-
- Schauvliege R., Janssens S., Beyaert R. (2006) Pellino proteins are more than scaffold proteins in TLR/IL-1R signalling: a role as novel RING E3-ubiquitin-ligases. FEBS Lett. 580, 4697–4702 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

