OsbZIP58, a basic leucine zipper transcription factor, regulates starch biosynthesis in rice endosperm
- PMID: 23846875
- PMCID: PMC3733163
- DOI: 10.1093/jxb/ert187
OsbZIP58, a basic leucine zipper transcription factor, regulates starch biosynthesis in rice endosperm
Abstract
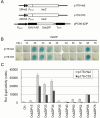
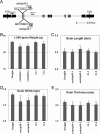
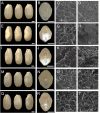
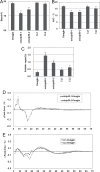
Starch composition and the amount in endosperm, both of which contribute dramatically to seed yield, cooking quality, and taste in cereals, are determined by a series of complex biochemical reactions. However, the mechanism regulating starch biosynthesis in cereal seeds is not well understood. This study showed that OsbZIP58, a bZIP transcription factor, is a key transcriptional regulator controlling starch synthesis in rice endosperm. OsbZIP58 was expressed mainly in endosperm during active starch synthesis. osbzip58 null mutants displayed abnormal seed morphology with altered starch accumulation in the white belly region and decreased amounts of total starch and amylose. Moreover, osbzip58 had a higher proportion of short chains and a lower proportion of intermediate chains of amylopectin. Furthermore, OsbZIP58 was shown to bind directly to the promoters of six starch-synthesizing genes, OsAGPL3, Wx, OsSSIIa, SBE1, OsBEIIb, and ISA2, and to regulate their expression. These findings indicate that OsbZIP58 functions as a key regulator of starch synthesis in rice seeds and provide new insights into seed quality control.
Keywords: Endosperm; OsbZIP58; coordination; rice; starch biosynthesis..
Figures

Similar articles
-
OPAQUE3, encoding a transmembrane bZIP transcription factor, regulates endosperm storage protein and starch biosynthesis in rice.Plant Commun. 2022 Nov 14;3(6):100463. doi: 10.1016/j.xplc.2022.100463. Epub 2022 Oct 18. Plant Commun. 2022. PMID: 36258666 Free PMC article.
-
High temperature inhibits the accumulation of storage materials by inducing alternative splicing of OsbZIP58 during filling stage in rice.Plant Cell Environ. 2020 Aug;43(8):1879-1896. doi: 10.1111/pce.13779. Epub 2020 Jun 9. Plant Cell Environ. 2020. PMID: 32335936
-
Overexpression of ZmbZIP22 gene alters endosperm starch content and composition in maize and rice.Plant Sci. 2019 Jun;283:407-415. doi: 10.1016/j.plantsci.2019.03.001. Epub 2019 Mar 5. Plant Sci. 2019. PMID: 31128711
-
Starch biosynthesis in cereal endosperm.Plant Physiol Biochem. 2010 Jun;48(6):383-92. doi: 10.1016/j.plaphy.2010.03.006. Epub 2010 Mar 24. Plant Physiol Biochem. 2010. PMID: 20400324 Review.
-
Proteomics and Post-Translational Modifications of Starch Biosynthesis-Related Proteins in Developing Seeds of Rice.Int J Mol Sci. 2021 May 31;22(11):5901. doi: 10.3390/ijms22115901. Int J Mol Sci. 2021. PMID: 34072759 Free PMC article. Review.
Cited by
-
OPAQUE3, encoding a transmembrane bZIP transcription factor, regulates endosperm storage protein and starch biosynthesis in rice.Plant Commun. 2022 Nov 14;3(6):100463. doi: 10.1016/j.xplc.2022.100463. Epub 2022 Oct 18. Plant Commun. 2022. PMID: 36258666 Free PMC article.
-
Functional Analysis of Starch Metabolism in Plants.Plants (Basel). 2020 Sep 6;9(9):1152. doi: 10.3390/plants9091152. Plants (Basel). 2020. PMID: 32899939 Free PMC article.
-
Starch Components, Starch Properties and Appearance Quality of Opaque Kernels from Rice Mutants.Molecules. 2019 Dec 13;24(24):4580. doi: 10.3390/molecules24244580. Molecules. 2019. PMID: 31847303 Free PMC article.
-
High Daytime Temperature Responsive MicroRNA Profiles in Developing Grains of Rice Varieties with Contrasting Chalkiness.Int J Mol Sci. 2023 Jul 19;24(14):11631. doi: 10.3390/ijms241411631. Int J Mol Sci. 2023. PMID: 37511395 Free PMC article.
-
A review of starch biosynthesis in cereal crops and its potential breeding applications in rice (Oryza Sativa L.).PeerJ. 2021 Dec 22;9:e12678. doi: 10.7717/peerj.12678. eCollection 2021. PeerJ. 2021. PMID: 35036154 Free PMC article.
References
-
- Burrell MM. 2003. Starch: the need for improved quality or quantity--an overview. Journal of Experimental Botany 54, 451–456 - PubMed
-
- Cai Y, Xie DL, Wang ZY, Hong MM. 2002. Interaction of rice bZIP protein REB with the 5′-upstream region of both rice sbe1 gene and waxy gene. Chinese Science Bulletin 47, 310–314
-
- Chen L, Wang ZY, Zhang JL, Cai XL, Hong MM. 1996a. Nuclear protein binding site in 5ʹ-upstream region of the rice waxy gene. Acta Phytophysiologica Sinica (in Chinese) 22, 349–356
-
- Chen L, Xing YY, Zhang JL, Wang ZY, Hong MM. 1996b. Isolation of the genes encording DNA bingding protein Interacting with 5ʹ upsteam region of the rice waxy gene by using yeast one-hybrid system. Acta Phytophysiologica Sinica 25, 110–114 (in Chinese).
-
- Cheng SJ, Wang ZY, Hong MM. 2002. Rice bZIP protein, REB, interacts with GCN4 motif in promoter of Waxy gene. Science China Life Sciences 45, 352–360 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

