Identification of an overprinting gene in Merkel cell polyomavirus provides evolutionary insight into the birth of viral genes
- PMID: 23847207
- PMCID: PMC3732942
- DOI: 10.1073/pnas.1303526110
Identification of an overprinting gene in Merkel cell polyomavirus provides evolutionary insight into the birth of viral genes
Abstract
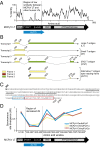
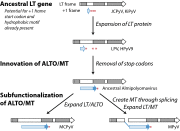
Many viruses use overprinting (alternate reading frame utilization) as a means to increase protein diversity in genomes severely constrained by size. However, the evolutionary steps that facilitate the de novo generation of a novel protein within an ancestral ORF have remained poorly characterized. Here, we describe the identification of an overprinting gene, expressed from an Alternate frame of the Large T Open reading frame (ALTO) in the early region of Merkel cell polyomavirus (MCPyV), the causative agent of most Merkel cell carcinomas. ALTO is expressed during, but not required for, replication of the MCPyV genome. Phylogenetic analysis reveals that ALTO is evolutionarily related to the middle T antigen of murine polyomavirus despite almost no sequence similarity. ALTO/MT arose de novo by overprinting of the second exon of T antigen in the common ancestor of a large clade of mammalian polyomaviruses. Taking advantage of the low evolutionary divergence and diverse sampling of polyomaviruses, we propose evolutionary transitions that likely gave birth to this protein. We suggest that two highly constrained regions of the large T antigen ORF provided a start codon and C-terminal hydrophobic motif necessary for cellular localization of ALTO. These two key features, together with stochastic erasure of intervening stop codons, resulted in a unique protein-coding capacity that has been preserved ever since its birth. Our study not only reveals a previously undefined protein encoded by several polyomaviruses including MCPyV, but also provides insight into de novo protein evolution.
Keywords: disordered motifs; gene evolution; synonymous substitution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

