Phylogenetic diversity and functional gene patterns of sulfur-oxidizing subseafloor Epsilonproteobacteria in diffuse hydrothermal vent fluids
- PMID: 23847608
- PMCID: PMC3703533
- DOI: 10.3389/fmicb.2013.00185
Phylogenetic diversity and functional gene patterns of sulfur-oxidizing subseafloor Epsilonproteobacteria in diffuse hydrothermal vent fluids
Abstract
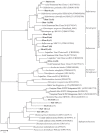
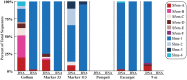
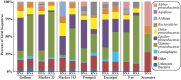
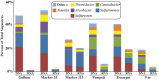
Microorganisms throughout the dark ocean use reduced sulfur compounds for chemolithoautotrophy. In many deep-sea hydrothermal vents, sulfide oxidation is quantitatively the most important chemical energy source for microbial metabolism both at and beneath the seafloor. In this study, the presence and activity of vent endemic Epsilonproteobacteria was examined in six low-temperature diffuse vents over a range of geochemical gradients from Axial Seamount, a deep-sea volcano in the Northeast Pacific. PCR primers were developed and applied to target the sulfur oxidation soxB gene of Epsilonproteobacteria. soxB genes belonging to the genera Sulfurimonas and Sulfurovum are both present and expressed at most diffuse vent sites, but not in background seawater. Although Sulfurovum-like soxB genes were detected in all fluid samples, the RNA profiles were nearly identical among the vents and suggest that Sulfurimonas-like species are the primary Epsilonproteobacteria responsible for actively oxidizing sulfur via the Sox pathway at each vent. Community patterns of subseafloor Epsilonproteobacteria 16S rRNA genes were best matched to methane concentrations in vent fluids, as well as individual vent locations, indicating that both geochemistry and geographical isolation play a role in structuring subseafloor microbial populations. The data show that in the subseafloor at Axial Seamount, Epsilonproteobacteria are expressing the soxB gene and that microbial patterns in community distribution are linked to both vent location and chemistry.
Keywords: 16S rRNA; Epsilonproteobacteria; functional genes; hydrothermal vent microbiology; subseafloor; sulfur oxidation.
Figures
References
-
- Bonch-Osmolovskaya E. A., Perevalova A. A., Kolganova T. V, Rusanov I. I., Jeanthon C., Pimenov N. V. (2011). Activity and distribution of thermophilic prokaryotes in hydrothermal fluid, sulfidic structures, and sheaths of Alvinellids (East Pacific Rise, 13N). Appl. Environ. Microbiol. 77, 2803–2806 10.1128/AEM.02266-10 - DOI - PMC - PubMed
-
- Bourbonnais A., Juniper S. K., Butterfield D. A., Devol A. H., Kuypers M. M. M., Lavik G., et al. (2012). Activity and abundance of denitrifying bacteria in the subsurface biosphere of diffuse hydrothermal vents of the Juan de Fuca Ridge. Biogeosciences 9, 4661–4678 10.5194/bg-9-4661-2012 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

