Principal component analysis reveals the 1000 Genomes Project does not sufficiently cover the human genetic diversity in Asia
- PMID: 23847652
- PMCID: PMC3701331
- DOI: 10.3389/fgene.2013.00127
Principal component analysis reveals the 1000 Genomes Project does not sufficiently cover the human genetic diversity in Asia
Abstract
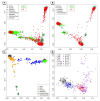
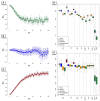
The 1000 Genomes Project (1KG) aims to provide a comprehensive resource on human genetic variations. With an effort of sequencing 2,500 individuals, 1KG is expected to cover the majority of the human genetic diversities worldwide. In this study, using analysis of population structure based on genome-wide single nucleotide polymorphisms (SNPs) data, we examined and evaluated the coverage of genetic diversity of 1KG samples with the available genome-wide SNP data of 3,831 individuals representing 140 population samples worldwide. We developed a method to quantitatively measure and evaluate the genetic diversity revealed by population structure analysis. Our results showed that the 1KG does not have sufficient coverage of the human genetic diversity in Asia, especially in Southeast Asia. We suggested a good coverage of Southeast Asian populations be considered in 1KG or a regional effort be initialized to provide a more comprehensive characterization of the human genetic diversity in Asia, which is important for both evolutionary and medical studies in the future.
Keywords: 1000 Genomes Project; Human Genome Diversity Project; Pan-Asian SNP Project; human genetic diversity; population structure; principal component analysis; single nucleotide polymorphisms.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources

