Detection of Human Endogenous Retrovirus K (HERV-K) Transcripts in Human Prostate Cancer Cell Lines
- PMID: 23847768
- PMCID: PMC3705622
- DOI: 10.3389/fonc.2013.00180
Detection of Human Endogenous Retrovirus K (HERV-K) Transcripts in Human Prostate Cancer Cell Lines
Abstract
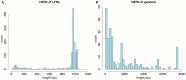
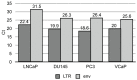
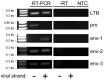
Human endogenous retroviruses (HERVs) are transcribed in many cancers including prostate cancer. Human endogenous retrovirus K (HERV-K) of the HML2 subtype is the most recently integrated and most intact retrovirus in the human genome, with many of the viral genomes encoding full- or partial-length viral proteins. To assess transcripts of HERV-K in prostate cancer cell lines and identify the specific HERV-K elements in the human genome that are transcribed, reverse transcriptase-PCR (RT-PCR) and cDNA sequencing were undertaken. Strand-specific RT-PCR, plasmid subcloning, and cDNA sequencing detected the presence of HERV-K(HML2) coding strand transcripts within four prostate cell lines (LNCaP, DU145, PC3, and VCaP). RT-PCR across splice junctions revealed splicing variants for env gene mRNA in three cell lines, two involving previously undescribed alternative splice sites. To determine the HERV-K loci from which the transcripts arose, RepeatMasker was used to compile a list of over 200 HERV-K internal genome segment fragments and over 1,000 HERV-K solo long terminal repeat (LTR) fragments in the human genome. Surprisingly, the sequences identified from internal positions of the viral genome were mostly smaller segments, while the LTRs were relatively intact. Possible reasons for this are discussed. The transcripts in the cell lines tested, arose from several HERV-K loci, with some proviruses being detected in multiple cell lines and others in only one of the four used. In some instances, transcripts from viral antisense strands was also detected. In addition, transcripts from both strands of solo LTRs were detected. These data show that transcripts from HERV-K loci commonly occur in prostate cancer cell lines and that transcription of either strand can occur. They also emphasize the importance of single nucleotide level analysis to identify the specific, individual HERV-K loci that are transcribed, and indicate that HERV-K expression in prostate cancer warrants further study.
Keywords: HERV-K; RT-PCR; cancer; endogenous retroviruses; evolution; prostate cancer; unconventional splicing.
Figures



References
-
- Belshaw R, Dawson ALA, Woolven-Allen J, Redding J, Burt A, Tristem M. Genomewide screening reveals high levels of insertional polymorphism in the human endogenous retrovirus family HERV-K(HML2): implications for present-day activity. J Virol (2005) 79:12507–1410.1128/JVI.79.19.12507-12514.2005 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

