Structural basis for the divergence of substrate specificity and biological function within HAD phosphatases in lipopolysaccharide and sialic acid biosynthesis
- PMID: 23848398
- PMCID: PMC3966652
- DOI: 10.1021/bi400659k
Structural basis for the divergence of substrate specificity and biological function within HAD phosphatases in lipopolysaccharide and sialic acid biosynthesis
Abstract
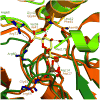
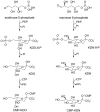
The haloacid dehalogenase enzyme superfamily (HADSF) is largely composed of phosphatases that have been particularly successful at adaptating to novel biological functions relative to members of other phosphatase families. Herein, we examine the structural basis for the divergence of function in two bacterial homologues: 2-keto-3-deoxy-d-manno-octulosonate 8-phosphate phosphohydrolase (KDO8P phosphatase, KDO8PP) and 2-keto-3-deoxy-9-O-phosphonononic acid phosphohydrolase (KDN9P phosphatase, KDN9PP). KDO8PP and KDN9PP catalyze the final step in KDO and KDN synthesis, respectively, prior to transfer to CMP to form the activated sugar nucleotide. KDO8PP and KDN9PP orthologs derived from an evolutionarily diverse collection of bacterial species were subjected to steady-state kinetic analysis to determine their specificities toward catalyzed KDO8P and KDN9P hydrolysis. Although each enzyme was more active with its biological substrate, the degree of selectivity (as defined by the ratio of kcat/Km for KDO8P vs KDN9P) varied significantly. High-resolution X-ray structure determination of Haemophilus influenzae KDO8PP bound to KDO/VO3(-) and Bacteriodes thetaiotaomicron KDN9PP bound to KDN/VO3(-) revealed the substrate-binding residues. The structures of the KDO8PP and KDN9PP orthologs were also determined to reveal the differences in their active-site structures that underlie the variation in substrate preference. Bioinformatic analysis was carried out to define the sequence divergence among KDN9PP and KDO8PP orthologs. The KDN9PP orthologs were found to exist as single-domain proteins or fused with the pathway nucleotidyl transferases; the fusion of KDO8PP with the transferase is rare. The KDO8PP and KDN9PP orthologs share a stringently conserved Arg residue that forms a salt bridge with the substrate carboxylate group. The split of the KDN9PP lineage from the KDO8PP orthologs is easily tracked by the acquisition of a Glu/Lys pair that supports KDN9P binding. Moreover, independently evolved lineages of KDO8PP orthologs exist, and are separated by diffuse active-site sequence boundaries. We infer a high tolerance of the KDO8PP catalytic platform to amino acid replacements that in turn influence substrate specificity changes and thereby facilitate the divergence in biological function.
Figures





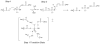
References
-
- Rangarajan ES, Proteau A, Wagner J, Hung MN, Matte A, Cygler M. Structural snapshots of Escherichia coli histidinol phosphate phosphatase along the reaction pathway. J Biol Chem. 2006;281:37930–37941. - PubMed
-
- Burroughs AM, Allen KN, Dunaway-Mariano D, Aravind L. Evolutionary genomics of the HAD superfamily: understanding the structural adaptations and catalytic diversity in a superfamily of phosphoesterases and allied enzymes. J Mol Biol. 2006;361:1003–1034. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

