The papillomavirus E2 proteins
- PMID: 23849793
- PMCID: PMC3783563
- DOI: 10.1016/j.virol.2013.06.006
The papillomavirus E2 proteins
Abstract
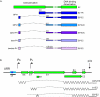
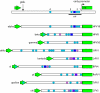
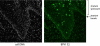
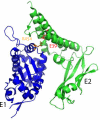
The papillomavirus E2 proteins are pivotal to the viral life cycle and have well characterized functions in transcriptional regulation, initiation of DNA replication and partitioning the viral genome. The E2 proteins also function in vegetative DNA replication, post-transcriptional processes and possibly packaging. This review describes structural and functional aspects of the E2 proteins and their binding sites on the viral genome. It is intended to be a reference guide to this viral protein.
Keywords: E2; Genomics; HPV; Mutation; Papillomavirus; Replication; Structure; Tethering; Transcription.
Published by Elsevier Inc.
Figures
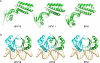






References
-
- Abbate EA, Voitenleitner C, Botchan MR. Structure of the papillomavirus DNA-tethering complex E2:Brd4 and a peptide that ablates HPV chromosomal association. Mol.Cell. 2006;24:877–889. - PubMed
-
- Akgul B, Pfefferle R, Marcuzzi GP, Zigrino P, Krieg T, Pfister H, Mauch C. Expression of matrix metalloproteinase (MMP)-2, MMP-9, MMP-13, and MT1-MMP in skin tumors of human papillomavirus type 8 transgenic mice. Exp.Dermatol. 2006;15:35–42. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

