LuxR- and luxI-type quorum-sensing circuits are prevalent in members of the Populus deltoides microbiome
- PMID: 23851092
- PMCID: PMC3754149
- DOI: 10.1128/AEM.01417-13
LuxR- and luxI-type quorum-sensing circuits are prevalent in members of the Populus deltoides microbiome
Abstract
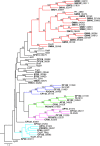
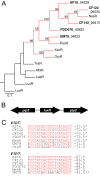
We are interested in the root microbiome of the fast-growing Eastern cottonwood tree, Populus deltoides. There is a large bank of bacterial isolates from P. deltoides, and there are 44 draft genomes of bacterial endophyte and rhizosphere isolates. As a first step in efforts to understand the roles of bacterial communication and plant-bacterial signaling in P. deltoides, we focused on the prevalence of acyl-homoserine lactone (AHL) quorum-sensing-signal production and reception in members of the P. deltoides microbiome. We screened 129 bacterial isolates for AHL production using a broad-spectrum bioassay that responds to many but not all AHLs, and we queried the available genome sequences of microbiome isolates for homologs of AHL synthase and receptor genes. AHL signal production was detected in 40% of 129 strains tested. Positive isolates included members of the Alpha-, Beta-, and Gammaproteobacteria. Members of the luxI family of AHL synthases were identified in 18 of 39 proteobacterial genomes, including genomes of some isolates that tested negative in the bioassay. Members of the luxR family of transcription factors, which includes AHL-responsive factors, were more abundant than luxI homologs. There were 72 in the 39 proteobacterial genomes. Some of the luxR homologs appear to be members of a subfamily of LuxRs that respond to as-yet-unknown plant signals rather than bacterial AHLs. Apparently, there is a substantial capacity for AHL cell-to-cell communication in proteobacteria of the P. deltoides microbiota, and there are also Proteobacteria with LuxR homologs of the type hypothesized to respond to plant signals or cues.
Figures

References
-
- Case RJ, Labbate M, Kjelleberg S. 2008. AHL-driven quorum-sensing circuits: their frequency and function among the proteobacteria. ISME J. 2:345–349 - PubMed
-
- Waters CM, Bassler BL. 2005. Quorum sensing: cell-to-cell communication. Annu. Rev. Cell Dev. Biol. 21:19–46 - PubMed
-
- Whitehead NA, Barnard AM, Slater H, Simpson NJ, Salmond GP. 2001. Quorum-sensing in gram-negative bacteria. FEMS Microbial Rev. 25:365–404 - PubMed
-
- Schaefer AL, Greenberg EP, Oliver CM, Oda Y, Huang JJ, Bittan-Banin G, Peres CM, Schmidt S, Juhaszova K, Sufrin JR, Harwood CS. 2008. A new class of homoserine lactone quorum-sensing signals. Nature 454:595–599 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

