Specific evidence of low-dimensional continuous attractor dynamics in grid cells
- PMID: 23852111
- PMCID: PMC3797513
- DOI: 10.1038/nn.3450
Specific evidence of low-dimensional continuous attractor dynamics in grid cells
Abstract
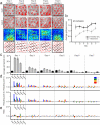
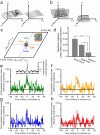
We examined simultaneously recorded spikes from multiple rat grid cells, to explain mechanisms underlying their activity. Among grid cells with similar spatial periods, the population activity was confined to lie close to a two-dimensional (2D) manifold: grid cells differed only along two dimensions of their responses and otherwise were nearly identical. Relationships between cell pairs were conserved despite extensive deformations of single-neuron responses. Results from novel environments suggest such structure is not inherited from hippocampal or external sensory inputs. Across conditions, cell-cell relationships are better conserved than responses of single cells. Finally, the system is continually subject to perturbations that, were the 2D manifold not attractive, would drive the system to inhabit a different region of state space than observed. These findings have strong implications for theories of grid-cell activity and substantiate the general hypothesis that the brain computes using low-dimensional continuous attractors.
Figures





References
-
- Seung HS, Lee D. The manifold ways of perception. Science. 2000;290:2268–2269. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

