The miniature genome of a carnivorous plant Genlisea aurea contains a low number of genes and short non-coding sequences
- PMID: 23855885
- PMCID: PMC3728226
- DOI: 10.1186/1471-2164-14-476
The miniature genome of a carnivorous plant Genlisea aurea contains a low number of genes and short non-coding sequences
Abstract
Background: Genlisea aurea (Lentibulariaceae) is a carnivorous plant with unusually small genome size - 63.6 Mb - one of the smallest known among higher plants. Data on the genome sizes and the phylogeny of Genlisea suggest that this is a derived state within the genus. Thus, G. aurea is an excellent model organism for studying evolutionary mechanisms of genome contraction.
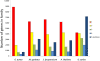
Results: Here we report sequencing and de novo draft assembly of G. aurea genome. The assembly consists of 10,687 contigs of the total length of 43.4 Mb and includes 17,755 complete and partial protein-coding genes. Its comparison with the genome of Mimulus guttatus, another representative of higher core Lamiales clade, reveals striking differences in gene content and length of non-coding regions.
Conclusions: Genome contraction was a complex process, which involved gene loss and reduction of lengths of introns and intergenic regions, but not intron loss. The gene loss is more frequent for the genes that belong to multigenic families indicating that genetic redundancy is an important prerequisite for genome size reduction.
Figures
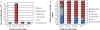
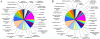
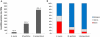

References
-
- Pellicer J, Fay MF, Leitch IJ. The largest eukaryotic genome of them all? Bot J Linn Soc. 2010;164:10–15. doi: 10.1111/j.1095-8339.2010.01072.x. - DOI
-
- Derelle E, Ferraz C, Rombauts S, Rouzé P, Worden AZ, Robbens S, Partensky F, Degroeve S, Echeynié S, Cooke R, Saeys Y, Wuyts J, Jabbari K, Bowler C, Panaud O, Piégu B, Ball SG, Ral J-P, Bouget F-Y, Piganeau G, De Baets B, Picard A, Delseny M, Demaille J, Van de Peer Y, Moreau H. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc Natl Acad Sci USA. 2006;103:11647–11652. doi: 10.1073/pnas.0604795103. - DOI - PMC - PubMed
-
- Town CD, Cheung F, Maiti R, Crabtree J, Haas BJ, Wortman JR, Hine EE, Althoff R, Arbogast TS, Tallon LJ, Vigouroux M, Trick M, Bancroft I. Comparative genomics of Brassica oleracea and Arabidopsis thaliana reveal gene loss, fragmentation, and dispersal after polyploidy. Plant Cell. 2006;18:1348–1359. doi: 10.1105/tpc.106.041665. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

