Topological and phylogenetic analyses of bacterial holin families and superfamilies
- PMID: 23856191
- PMCID: PMC3788059
- DOI: 10.1016/j.bbamem.2013.07.004
Topological and phylogenetic analyses of bacterial holin families and superfamilies
Abstract
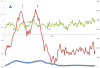
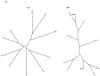
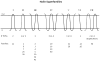
Holins are small "hole-forming" transmembrane proteins that mediate bacterial cell lysis during programmed cell death or following phage infection. We have identified fifty two families of established or putative holins and have included representative members of these proteins in the Transporter Classification Database (TCDB; www.tcdb.org). We have identified the organismal sources of members of these families, calculated their average protein sizes, estimated their topologies and determined their relative family sizes. Topological analyses suggest that these proteins can have 1, 2, 3 or 4 transmembrane α-helical segments (TMSs), and members of a single family are frequently, but not always, of a single topology. In one case, proteins of a family proved to have either 2 or 4 TMSs, and the latter arose by intragenic duplication of a primordial 2 TMS protein-encoding gene resembling the former. Using established statistical approaches, some of these families have been shown to be related by common descent. Seven superfamilies, including 21 of the 52 recognized families were identified. Conserved motif and Pfam analyses confirmed most superfamily assignments. These results serve to expand upon the scope of channel-forming bacterial holins.
Keywords: Autolysin; Holin; Phylogeny; Superfamily; Transmembrane pore; “Hole-forming”.
© 2013.
Figures














References
-
- Young R, Blasi U. Holins: form and function in bacteriophage lysis. FEMS Microbiol Rev. 1995;17:191–205. - PubMed
-
- Saier MH, Ma CH, Rodgers L, Tamang DG, Yen MR. Protein secretion and membrane insertion systems in bacteria and eukaryotic organelles. Adv Appl Microbiol. 2008;65:141–197. - PubMed
-
- Wang IN, Smith DL, Young R. Holins: the protein clocks of bacteriophage infections. Annu Rev Microbiol. 2000;54:799–825. - PubMed
-
- Desvaux M, Hebraud M, Talon R, Henderson IR. Secretion and subcellular localizations of bacterial proteins: a semantic awareness issue. Trends Microbiol. 2009;17:139–145. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

