The presence, role and clinical use of spermatozoal RNAs
- PMID: 23856356
- PMCID: PMC3796946
- DOI: 10.1093/humupd/dmt031
The presence, role and clinical use of spermatozoal RNAs
Abstract
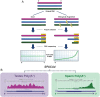
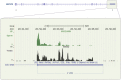
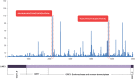
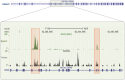
BACKGROUND Spermatozoa are highly differentiated, transcriptionally inert cells characterized by a compact nucleus with minimal cytoplasm. Nevertheless they contain a suite of unique RNAs that are delivered to oocyte upon fertilization. They are likely integrated as part of many different processes including genome recognition, consolidation-confrontation, early embryonic development and epigenetic transgenerational inherence. Spermatozoal RNAs also provide a window into the developmental history of each sperm thereby providing biomarkers of fertility and pregnancy outcome which are being intensely studied. METHODS Literature searches were performed to review the majority of spermatozoal RNA studies that described potential functions and clinical applications with emphasis on Next-Generation Sequencing. Human, mouse, bovine and stallion were compared as their distribution and composition of spermatozoal RNAs, using these techniques, have been described. RESULTS Comparisons highlighted the complexity of the population of spermatozoal RNAs that comprises rRNA, mRNA and both large and small non-coding RNAs. RNA-seq analysis has revealed that only a fraction of the larger RNAs retain their structure. While rRNAs are the most abundant and are highly fragmented, ensuring a translationally quiescent state, other RNAs including some mRNAs retain their functional potential, thereby increasing the opportunity for regulatory interactions. Abundant small non-coding RNAs retained in spermatozoa include miRNAs and piRNAs. Some, like miR-34c are essential to the early embryo development required for the first cellular division. Others like the piRNAs are likely part of the genomic dance of confrontation and consolidation. Other non-coding spermatozoal RNAs include transposable elements, annotated lnc-RNAs, intronic retained elements, exonic elements, chromatin-associated RNAs, small-nuclear ILF3/NF30 associated RNAs, quiescent RNAs, mse-tRNAs and YRNAs. Some non-coding RNAs are known to act as epigenetic modifiers, inducing histone modifications and DNA methylation, perhaps playing a role in transgenerational epigenetic inherence. Transcript profiling holds considerable potential for the discovery of fertility biomarkers for both agriculture and human medicine. Comparing the differential RNA profiles of infertile and fertile individuals as well as assessing species similarities, should resolve the regulatory pathways contributing to male factor infertility. CONCLUSIONS Dad delivers a complex population of RNAs to the oocyte at fertilization that likely influences fertilization, embryo development, the phenotype of the offspring and possibly future generations. Development is continuing on the use of spermatozoal RNA profiles as phenotypic markers of male factor status for use as clinical diagnostics of the father's contribution to the birth of a healthy child.
Keywords: embryogenesis; epigenetics modifiers; fertility biomarkers; spermatozoal RNA; transgenerational epigenetic inherence.
Figures




References
-
- Adams GP, Pierson RA. Bovine model for study of ovarian follicular dynamics in humans. Theriogenology. 1995;43:113–120.
-
- Anton E, Krawetz SA. Spermatozoa as biomarkers for the assessment of human male infertility and genotoxicity. Syst Biol Reprod Med. 2012;58:41–50. - PubMed
-
- Arangasamy A, Kasimanickam VR, DeJarnette JM, Kasimanickam RK. Association of CRISP2, CCT8, PEBP1 mRNA abundance in sperm and sire conception rate in Holstein bulls. Theriogenology. 2011;76:570–577. - PubMed
-
- Aravin AA, Hannon GJ. Small RNA silencing pathways in germ and stem cells. Cold Spring Harb Symp Quant Biol. 2008;73:283–290. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

