Pig transgenesis by piggyBac transposition in combination with somatic cell nuclear transfer
- PMID: 23857557
- PMCID: PMC3838457
- DOI: 10.1007/s11248-013-9729-0
Pig transgenesis by piggyBac transposition in combination with somatic cell nuclear transfer
Abstract
The production of animals by somatic cell nuclear transfer (SCNT) is inefficient, with approximately 2% of micromanipulated oocytes going to term and resulting in live births. However, it is the most commonly used method for the generation of cloned transgenic livestock as it facilitates the attainment of transgenic animals once the nuclear donor cells are stably transfected and more importantly as alternatives methods of transgenesis in farm animals have proven even less efficient. Here we describe piggyBac-mediated transposition of a transgene into porcine primary cells and use of these genetically modified cells as nuclear donors for the generation of transgenic pigs by SCNT. Gene transfer by piggyBac transposition serves to provide an alternative approach for the transfection of nuclear donor cells used in SCNT.
Figures
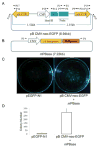
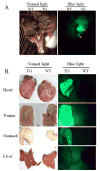
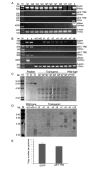

References
-
- Al-Mashhadi RH, Sorensen CB, Kragh PM, Christoffersen C, Mortensen MB, Tolbod LP, Thim T, Du Y, Li J, Liu Y, Moldt B, Schmidt M, Vajta G, Larsen T, Purup S, Bolund L, Nielsen LB, Callesen H, Falk E, Mikkelsen JG, Bentzon JF. Familial hypercholesterolemia and atherosclerosis in cloned minipigs created by DNA transposition of a human PCSK9 gain-of-function mutant. Science translational medicine. 2013;5(166):166ra–161. doi: 10.1126/scitranslmed.3004853. - DOI - PubMed
-
- Betthauser J, Forsberg E, Augenstein M, Childs L, Eilertsen K, Enos J, Forsythe T, Golueke P, Jurgella G, Koppang R, Lesmeister T, Mallon K, Mell G, Misica P, Pace M, Pfister-Genskow M, Strelchenko N, Voelker G, Watt S, Thompson S, Bishop M. Production of cloned pigs from in vitro systems. Nature biotechnology. 2000;18(10):1055–1059. doi: 10.1038/80242. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

