Estrogen receptor β (ERβ1) transactivation is differentially modulated by the transcriptional coregulator Tip60 in a cis-acting element-dependent manner
- PMID: 23857583
- PMCID: PMC3757169
- DOI: 10.1074/jbc.M113.476952
Estrogen receptor β (ERβ1) transactivation is differentially modulated by the transcriptional coregulator Tip60 in a cis-acting element-dependent manner
Abstract
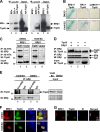
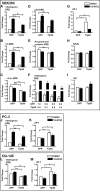
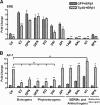
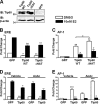
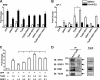
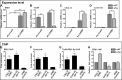
Estrogen receptor (ER) β1 and ERα have overlapping and distinct functions despite their common use of estradiol as the physiological ligand. These attributes are explained in part by their differential utilization of coregulators and ligands. Although Tip60 has been shown to interact with both receptors, its regulatory role in ERβ1 transactivation has not been defined. In this study, we found that Tip60 enhances transactivation of ERβ1 at the AP-1 site but suppresses its transcriptional activity at the estrogen-response element (ERE) site in an estradiol-independent manner. However, different estrogenic compounds can modify the Tip60 action. The corepressor activity of Tip60 at the ERE site is abolished by diarylpropionitrile, genistein, equol, and bisphenol A, whereas its coactivation at the AP-1 site is augmented by fulvestrant (ICI 182,780). GRIP1 is an important tethering mediator for ERs at the AP-1 site. We found that coexpression of GRIP1 synergizes the action of Tip60. Although Tip60 is a known acetyltransferase, it is unable to acetylate ERβ1, and its coregulatory functions are independent of its acetylation activity. In addition, we showed the co-occupancy of ERβ1 and Tip60 at ERE and AP-1 sites of ERβ1 target genes. Tip60 differentially regulates the endogenous expression of the target genes by modulating the binding of ERβ1 to the cis-regulatory regions. Thus, we have identified Tip60 as the first dual-function coregulator of ERβ1.
Keywords: AP-1; Acetyltransferase; Coregulator; ERβ1; Estrogen Receptor; Estrogen-response Element; Protein-Protein Interactions; Transcription; Transcription Enhancers; Transcription Repressor.
Figures


References
-
- Carroll J. S., Brown M. (2006) Estrogen receptor target gene: an evolving concept. Mol. Endocrinol. 20, 1707–1714 - PubMed
-
- Kushner P. J., Agard D. A., Greene G. L., Scanlan T. S., Shiau A. K., Uht R. M., Webb P. (2000) Estrogen receptor pathways to AP-1. J. Steroid Biochem. Mol. Biol. 74, 311–317 - PubMed
-
- Saville B., Wormke M., Wang F., Nguyen T., Enmark E., Kuiper G., Gustafsson J. A., Safe S. (2000) Ligand-, cell-, and estrogen receptor subtype (α/β)-dependent activation at GC-rich (Sp1) promoter elements. J. Biol. Chem. 275, 5379–5387 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01 ES020988/ES/NIEHS NIH HHS/United States
- P30 ES006096/ES/NIEHS NIH HHS/United States
- R01 CA015776/CA/NCI NIH HHS/United States
- P30ES006096/ES/NIEHS NIH HHS/United States
- I01 BX000675/BX/BLRD VA/United States
- R01 DK061084/DK/NIDDK NIH HHS/United States
- R01 ES015584/ES/NIEHS NIH HHS/United States
- U01ES019480/ES/NIEHS NIH HHS/United States
- R01CA015776/CA/NCI NIH HHS/United States
- R01DK061084/DK/NIDDK NIH HHS/United States
- U01ES020988/ES/NIEHS NIH HHS/United States
- R01ES015584/ES/NIEHS NIH HHS/United States
- U01 ES019480/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

