Immune history shapes specificity of pandemic H1N1 influenza antibody responses
- PMID: 23857983
- PMCID: PMC3727314
- DOI: 10.1084/jem.20130212
Immune history shapes specificity of pandemic H1N1 influenza antibody responses
Abstract
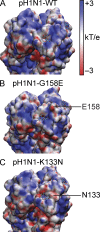
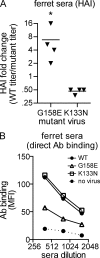
Human antibody responses against the 2009 pandemic H1N1 (pH1N1) virus are predominantly directed against conserved epitopes in the stalk and receptor-binding domain of the hemagglutinin (HA) protein. This is in stark contrast to pH1N1 antibody responses generated in ferrets, which are focused on the variable Sa antigenic site of HA. Here, we show that most humans born between 1983 and 1996 elicited pH1N1 antibody responses that are directed against an epitope near the HA receptor-binding domain. Importantly, most individuals born before 1983 or after 1996 did not elicit pH1N1 antibodies to this HA epitope. The HAs of most seasonal H1N1 (sH1N1) viruses that circulated between 1983 and 1996 possess a critical K133 amino acid in this HA epitope, whereas this amino acid is either mutated or deleted in most sH1N1 viruses circulating before 1983 or after 1996. We sequentially infected ferrets with a 1991 sH1N1 virus and then a pH1N1 virus. Sera isolated from these animals were directed against the HA epitope involving amino acid K133. These data suggest that the specificity of pH1N1 antibody responses can be shifted to epitopes near the HA receptor-binding domain after sequential infections with sH1N1 and pH1N1 viruses that share homology in this region.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

