Transcriptional dynamics of two seed compartments with opposing roles in Arabidopsis seed germination
- PMID: 23858430
- PMCID: PMC3762641
- DOI: 10.1104/pp.113.223511
Transcriptional dynamics of two seed compartments with opposing roles in Arabidopsis seed germination
Abstract
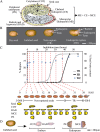
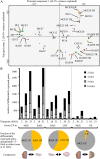
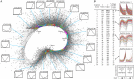
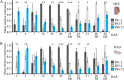
Seed germination is a critical stage in the plant life cycle and the first step toward successful plant establishment. Therefore, understanding germination is of important ecological and agronomical relevance. Previous research revealed that different seed compartments (testa, endosperm, and embryo) control germination, but little is known about the underlying spatial and temporal transcriptome changes that lead to seed germination. We analyzed genome-wide expression in germinating Arabidopsis (Arabidopsis thaliana) seeds with both temporal and spatial detail and provide Web-accessible visualizations of the data reported (vseed.nottingham.ac.uk). We show the potential of this high-resolution data set for the construction of meaningful coexpression networks, which provide insight into the genetic control of germination. The data set reveals two transcriptional phases during germination that are separated by testa rupture. The first phase is marked by large transcriptome changes as the seed switches from a dry, quiescent state to a hydrated and active state. At the end of this first transcriptional phase, the number of differentially expressed genes between consecutive time points drops. This increases again at testa rupture, the start of the second transcriptional phase. Transcriptome data indicate a role for mechano-induced signaling at this stage and subsequently highlight the fates of the endosperm and radicle: senescence and growth, respectively. Finally, using a phylotranscriptomic approach, we show that expression levels of evolutionarily young genes drop during the first transcriptional phase and increase during the second phase. Evolutionarily old genes show an opposite pattern, suggesting a more conserved transcriptome prior to the completion of germination.
Figures




References
-
- Braam J. (2005) In touch: plant responses to mechanical stimuli. New Phytol 165: 373–389 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

