An efficient method for long-term room temperature storage of RNA
- PMID: 23860045
- PMCID: PMC3925273
- DOI: 10.1038/ejhg.2013.145
An efficient method for long-term room temperature storage of RNA
Abstract
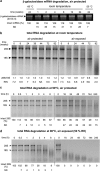
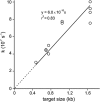
RNA is a tool used in many fields, from molecular and cellular biology to medicine and nanotechnology. For most of these uses, the integrity of RNA is required and must be maintained during storage. Even though freezing is currently the storage method of choice, the increasing number of samples to be stored and the costly use of a cold chain have highlighted the need for room temperature preservation methods. Here, we report a new room temperature technology that consists in drying RNA samples in the presence of a stabilizer in stainless steel minicapsules. These air- and water-tight capsules isolate RNA from the atmosphere and maintain an anhydrous and anoxic environment. Through the evaluation of RNA integrity over time at room temperature or 90 °C, we identified atmospheric humidity as a major deleterious factor. The degradation rate dependence in temperature fitted an Arrhenius model, with an activation energy of 28.5 kcal/mol and an extrapolated room temperature degradation rate of 3.2 10(-13)/nt/s (95% confidence interval: 2.3-4.2/nt/s). In these conditions, it is expected that an RNA molecule will be subjected to 0.7-1.3 cut every 1000 nucleotides per century. In addition, we showed that stored RNA is compatible for further analyses, such as reverse transcription-quantitative PCR. No significant change in the Cq values was observed over a simulated period of several decades. At last, our data are consistent with a sequence-independent degradation rate of RNA in the solid state.
Figures


References
-
- White HE, Matejtschuk P, Rigsby P, et al. Establishment of the first World Health Organization International Genetic Reference Panel for quantitation of BCR-ABL mRNA. Blood. 2010;116:e111–e117. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

