Peptidylarginine deiminases in citrullination, gene regulation, health and pathogenesis
- PMID: 23860259
- PMCID: PMC3775966
- DOI: 10.1016/j.bbagrm.2013.07.003
Peptidylarginine deiminases in citrullination, gene regulation, health and pathogenesis
Abstract
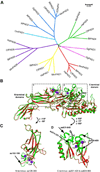
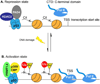
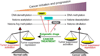
Peptidylarginine deiminases are a family of enzymes that mediate post-translational modifications of protein arginine residues by deimination or demethylimination to produce citrulline. In vitro, the activity of PADs is dependent on calcium and reductive reagents carrying a free sulfhydryl group. The discovery that PAD4 can target both arginine and methyl-arginine for citrullination about 10years ago renewed our interest in studying this family of enzymes in gene regulation and their physiological functions. The deregulation of PADs is involved in the etiology of multiple human diseases, including cancers and autoimmune disorders. There is a growing effort to develop isoform specific PAD inhibitors for disease treatment. However, the regulation of the activity of PADs in vivo remains largely elusive, and we expect that much will be learned about the role of these enzymes in a normal life cycle and under pathology conditions.
Keywords: Autoimmunity; Cancer; Gene regulation; Histone; Peptidylarginine deiminase.
© 2013.
Figures

References
-
- Rogers GE. Occurrence of Citrulline in Proteins. Nature. 1962;194:1149–1151. - PubMed
-
- Rogers G, Taylor L. The enzymic derivation of citrulline residues from arginine residues in situ during the biosynthesis of hair proteins that are cross-linked by isopeptide bonds. 1977 - PubMed
-
- Vossenaar ER, Zendman AJW, van Venrooij WJ, Pruijn GJM. PADa, growing family of citrullinating enzymes: genes, features and involvement in disease. BioEssays. 2003;25:1106–1118. - PubMed
-
- Baka Z, Gyorgy B, Geher P, Buzas EI, Falus A, Nagy G. Citrullination under physiological and pathological conditions. Joint, bone, spine : revue du rhumatisme. 2012;79:431–436. - PubMed
-
- Horibata S, Coonrod SA, Cherrington BD. Role for peptidylarginine deiminase enzymes in disease and female reproduction. The Journal of reproduction and development. 2012;58:274–282. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

