Assessing association between protein truncating variants and quantitative traits
- PMID: 23860716
- PMCID: PMC3777107
- DOI: 10.1093/bioinformatics/btt409
Assessing association between protein truncating variants and quantitative traits
Abstract
Motivation: In sequencing studies of common diseases and quantitative traits, power to test rare and low frequency variants individually is weak. To improve power, a common approach is to combine statistical evidence from several genetic variants in a region. Major challenges are how to do the combining and which statistical framework to use. General approaches for testing association between rare variants and quantitative traits include aggregating genotypes and trait values, referred to as 'collapsing', or using a score-based variance component test. However, little attention has been paid to alternative models tailored for protein truncating variants. Recent studies have highlighted the important role that protein truncating variants, commonly referred to as 'loss of function' variants, may have on disease susceptibility and quantitative levels of biomarkers. We propose a Bayesian modelling framework for the analysis of protein truncating variants and quantitative traits.
Results: Our simulation results show that our models have an advantage over the commonly used methods. We apply our models to sequence and exome-array data and discover strong evidence of association between low plasma triglyceride levels and protein truncating variants at APOC3 (Apolipoprotein C3).
Availability: Software is available from http://www.well.ox.ac.uk/~rivas/mamba
Figures
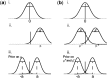
 ; (ii) distribution of the trait values under the alternative model,
; (ii) distribution of the trait values under the alternative model,  ; and (iii) 50:50 mixture of two normal distributions as prior for
; and (iii) 50:50 mixture of two normal distributions as prior for  . (b) Prior and sampling distribution for the GEM: (i) distribution of the trait values under the null model; (ii) under the alternative model, trait values are grouped around
. (b) Prior and sampling distribution for the GEM: (i) distribution of the trait values under the null model; (ii) under the alternative model, trait values are grouped around  and
and  ; and (iii) priors for
; and (iii) priors for  and
and 

 ) (Y-axis), for different values of n (written next to each point) the number of PTV carriers, for a fixed P-value of 0.001. The dashed line shows the density of the prior (restricted to positive values) on mean effect size under SEM (not on scale)
) (Y-axis), for different values of n (written next to each point) the number of PTV carriers, for a fixed P-value of 0.001. The dashed line shows the density of the prior (restricted to positive values) on mean effect size under SEM (not on scale)
References
-
- Chen WJ, et al. Exome sequencing identifies truncating mutations in PRRT2 that cause paroxysmal kinesigenic dyskinesia. Nat. Genet. 2011;43:1252–1255. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- RC2-HG005688/HG/NHGRI NIH HHS/United States
- 095552/Wellcome Trust/United Kingdom
- 083270/WT_/Wellcome Trust/United Kingdom
- R01 DK098032/DK/NIDDK NIH HHS/United States
- 085475/B/08/Z/WT_/Wellcome Trust/United Kingdom
- 076113/WT_/Wellcome Trust/United Kingdom
- 075491/Z/04/B/WT_/Wellcome Trust/United Kingdom
- RC2 DK088389/DK/NIDDK NIH HHS/United States
- 090532/Wellcome Trust/United Kingdom
- 086596/Z/08/Z/WT_/Wellcome Trust/United Kingdom
- 098381/Wellcome Trust/United Kingdom
- 085475/Z/08/Z/WT_/Wellcome Trust/United Kingdom
- G0601261/MRC_/Medical Research Council/United Kingdom
- 090367/WT_/Wellcome Trust/United Kingdom
- 095552/Z/11/Z/WT_/Wellcome Trust/United Kingdom
- 090532/Z/09/Z/WT_/Wellcome Trust/United Kingdom
- 09831/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

