Interpreting secondary cardiac disease variants in an exome cohort
- PMID: 23861362
- PMCID: PMC3887521
- DOI: 10.1161/CIRCGENETICS.113.000039
Interpreting secondary cardiac disease variants in an exome cohort
Abstract
Background: Massively parallel sequencing to identify rare variants is widely practiced in medical research and in the clinic. Genome and exome sequencing can identify the genetic cause of a disease (primary results), but it can also identify pathogenic variants underlying diseases that are not being sought (secondary or incidental results). A major controversy has developed surrounding the return of secondary results to research participants. We have piloted a method to analyze exomes to identify participants at risk for cardiac arrhythmias, cardiomyopathies, or sudden death.
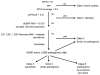
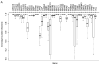
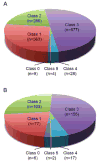
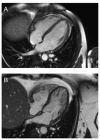
Methods and results: Exome sequencing was performed on 870 participants not selected for arrhythmia, cardiomyopathy, or a family history of sudden death. Exome data from 22 cardiac arrhythmia- and 41 cardiomyopathy-associated genes were analyzed using an algorithm that filtered results on genotype quality, frequency, and database information. We identified 1367 variants in the cardiomyopathy genes and 360 variants in the arrhythmia genes. Six participants had pathogenic variants associated with dilated cardiomyopathy (n=1), hypertrophic cardiomyopathy (n=2), left ventricular noncompaction (n=1), or long-QT syndrome (n=2). Two of these participants had evidence of cardiomyopathy and 1 had left ventricular noncompaction on echocardiogram. Three participants with likely pathogenic variants had prolonged QTc. Family history included unexplained sudden death among relatives.
Conclusions: Approximately 0.5% of participants in this study had pathogenic variants in known cardiomyopathy or arrhythmia genes. This high frequency may be due to self-selection, false positives, or underestimation of the prevalence of these conditions. We conclude that clinically important cardiomyopathy and dysrhythmia secondary variants can be identified in unselected exomes.
Keywords: arrhythmias, cardiac; cardiomyopathies; genetic susceptibility; genetic variation; genetics; genomics; long QT syndrome.
Conflict of interest statement
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

