Lariat sequencing in a unicellular yeast identifies regulated alternative splicing of exons that are evolutionarily conserved with humans
- PMID: 23861491
- PMCID: PMC3732940
- DOI: 10.1073/pnas.1218353110
Lariat sequencing in a unicellular yeast identifies regulated alternative splicing of exons that are evolutionarily conserved with humans
Abstract
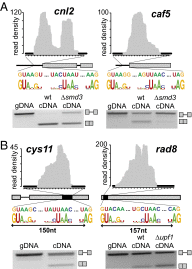
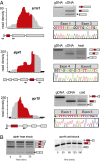
Alternative splicing is a potent regulator of gene expression that vastly increases proteomic diversity in multicellular eukaryotes and is associated with organismal complexity. Although alternative splicing is widespread in vertebrates, little is known about the evolutionary origins of this process, in part because of the absence of phylogenetically conserved events that cross major eukaryotic clades. Here we describe a lariat-sequencing approach, which offers high sensitivity for detecting splicing events, and its application to the unicellular fungus, Schizosaccharomyces pombe, an organism that shares many of the hallmarks of alternative splicing in mammalian systems but for which no previous examples of exon-skipping had been demonstrated. Over 200 previously unannotated splicing events were identified, including examples of regulated alternative splicing. Remarkably, an evolutionary analysis of four of the exons identified here as subject to skipping in S. pombe reveals high sequence conservation and perfect length conservation with their homologs in scores of plants, animals, and fungi. Moreover, alternative splicing of two of these exons have been documented in multiple vertebrate organisms, making these the first demonstrations of identical alternative-splicing patterns in species that are separated by over 1 billion y of evolution.
Keywords: phylogeny; post-transcriptional gene regulation; pre-mRNA splicing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Black DL. Protein diversity from alternative splicing: A challenge for bioinformatics and post-genome biology. Cell. 2000;103(3):367–370. - PubMed
-
- Lareau LF, Brooks AN, Soergel DAW, Meng Q, Brenner SE. The coupling of alternative splicing and nonsense-mediated mRNA decay. Adv Exp Med Biol. 2007;623:190–211. - PubMed
-
- McGlincy NJ, Smith CWJ. Alternative splicing resulting in nonsense-mediated mRNA decay: What is the meaning of nonsense? Trends Biochem Sci. 2008;33(8):385–393. - PubMed
-
- Breitbart RE, Andreadis A, Nadal-Ginard B. Alternative splicing: A ubiquitous mechanism for the generation of multiple protein isoforms from single genes. Annu Rev Biochem. 1987;56:467–495. - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

