Sequences encoding identical peptides for the analysis and manipulation of coding DNA
- PMID: 23861567
- PMCID: PMC3705626
- DOI: 10.6026/97320630009511
Sequences encoding identical peptides for the analysis and manipulation of coding DNA
Abstract
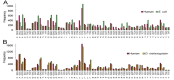
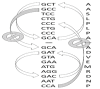
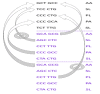
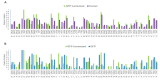
The use of sequences encoding identical peptides (SEIP) for the in silico analysis of coding DNA from different species has not been reported; the study of such sequences could directly reveal properties of coding DNA that are independent of peptide sequences. For practical purposes SEIP might also be manipulated for e.g. heterologous protein expression. We extracted 1,551 SEIP from human and E. coli and 2,631 SEIP from human and D. melanogaster. We then analyzed codon usage and intercodon dinucleotide tendencies and found differences in both, with more conspicuous disparities between human and E. coli than between human and D. melanogaster. We also briefly manipulated SEIP to find out if they could be used to create new coding sequences. We hence attempted replacement of human by E. coli codons via dicodon exchange but found that full replacement was not possible, this indicated robust species-specific dicodon tendencies. To test another form of codon replacement we isolated SEIP from human and the jellyfish green fluorescent protein (GFP) and we then re-constructed the GFP coding DNA with human tetra-peptide-coding sequences. Results provide proof-of-principle that SEIP may be used to reveal differences in the properties of coding DNA and to reconstruct in pieces a protein coding DNA with sequences from a different organism, the latter might be exploited in heterologous protein expression.
Keywords: codon allocation tendencies; codon pairs; green fluorescent protein; heterologous protein expression; intercodon dinucleotides; synonymous codons.
Figures



Similar articles
-
3-base periodicity in coding DNA is affected by intercodon dinucleotides.Bioinformation. 2011;6(9):327-9. doi: 10.6026/97320630006327. Epub 2011 Jul 19. Bioinformation. 2011. PMID: 21814388 Free PMC article.
-
Widespread position-specific conservation of synonymous rare codons within coding sequences.PLoS Comput Biol. 2017 May 5;13(5):e1005531. doi: 10.1371/journal.pcbi.1005531. eCollection 2017 May. PLoS Comput Biol. 2017. PMID: 28475588 Free PMC article.
-
Low-usage codons in Escherichia coli, yeast, fruit fly and primates.Gene. 1991 Aug 30;105(1):61-72. doi: 10.1016/0378-1119(91)90514-c. Gene. 1991. PMID: 1937008
-
A Code Within a Code: How Codons Fine-Tune Protein Folding in the Cell.Biochemistry (Mosc). 2021 Aug;86(8):976-991. doi: 10.1134/S0006297921080083. Biochemistry (Mosc). 2021. PMID: 34488574 Free PMC article. Review.
-
Analysis of synonymous codon usage patterns in the edible fungus Volvariella volvacea.Biotechnol Appl Biochem. 2017 Mar;64(2):218-224. doi: 10.1002/bab.1538. Epub 2016 Dec 15. Biotechnol Appl Biochem. 2017. PMID: 27696508 Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
