Engineering a more thermostable blue light photo receptor Bacillus subtilis YtvA LOV domain by a computer aided rational design method
- PMID: 23861663
- PMCID: PMC3701716
- DOI: 10.1371/journal.pcbi.1003129
Engineering a more thermostable blue light photo receptor Bacillus subtilis YtvA LOV domain by a computer aided rational design method
Abstract
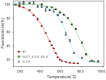
The ability to design thermostable proteins offers enormous potential for the development of novel protein bioreagents. In this work, a combined computational and experimental method was developed to increase the T m of the flavin mononucleotide based fluorescent protein Bacillus Subtilis YtvA LOV domain by 31 Celsius, thus extending its applicability in thermophilic systems. Briefly, the method includes five steps, the single mutant computer screening to identify thermostable mutant candidates, the experimental evaluation to confirm the positive selections, the computational redesign around the thermostable mutation regions, the experimental reevaluation and finally the multiple mutations combination. The adopted method is simple and effective, can be applied to other important proteins where other methods have difficulties, and therefore provides a new tool to improve protein thermostability.
Conflict of interest statement
I have read the journal's policy and have the following conflicts: A Chinese patent has been filed with part of the results in the paper.
Figures




References
-
- Steipe B, Schiller B, Pluckthun A, Steinbacher S (1994) SEQUENCE STATISTICS RELIABLY PREDICT STABILIZING MUTATIONS IN A PROTEIN DOMAIN. Journal of Molecular Biology 240: 188–192. - PubMed
-
- Mooers BHM, Datta D, Baase WA, Zollars ES, Mayo SL, et al. (2003) Repacking the core of T4 lysozyme by automated design. Journal of Molecular Biology 332: 741–756. - PubMed
-
- Blaber M, Lindstrom JD, Gassner N, Xu J, Dirk WH, et al. (1993) Energetic Cost and Structural Consequences of Burying a Hydroxyl Group within the Core of a Protein Determined from Ala-]Ser and Val-]Thr Substitutions in T4 Lysozyme. Biochemistry 32: 11363–11373. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

