Transcriptome analysis in sheepgrass (Leymus chinensis): a dominant perennial grass of the Eurasian Steppe
- PMID: 23861841
- PMCID: PMC3701641
- DOI: 10.1371/journal.pone.0067974
Transcriptome analysis in sheepgrass (Leymus chinensis): a dominant perennial grass of the Eurasian Steppe
Abstract
Background: Sheepgrass [Leymus chinensis (Trin.) Tzvel.] is an important perennial forage grass across the Eurasian Steppe and is known for its adaptability to various environmental conditions. However, insufficient data resources in public databases for sheepgrass limited our understanding of the mechanism of environmental adaptations, gene discovery and molecular marker development.
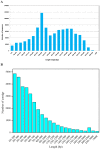
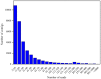
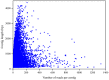
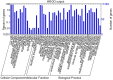
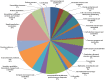
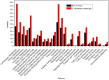
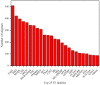
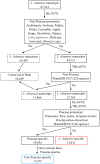
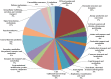
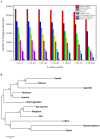
Results: The transcriptome of sheepgrass was sequenced using Roche 454 pyrosequencing technology. We assembled 952,328 high-quality reads into 87,214 unigenes, including 32,416 contigs and 54,798 singletons. There were 15,450 contigs over 500 bp in length. BLAST searches of our database against Swiss-Prot and NCBI non-redundant protein sequences (nr) databases resulted in the annotation of 54,584 (62.6%) of the unigenes. Gene Ontology (GO) analysis assigned 89,129 GO term annotations for 17,463 unigenes. We identified 11,675 core Poaceae-specific and 12,811 putative sheepgrass-specific unigenes by BLAST searches against all plant genome and transcriptome databases. A total of 2,979 specific freezing-responsive unigenes were found from this RNAseq dataset. We identified 3,818 EST-SSRs in 3,597 unigenes, and some SSRs contained unigenes that were also candidates for freezing-response genes. Characterizations of nucleotide repeats and dominant motifs of SSRs in sheepgrass were also performed. Similarity and phylogenetic analysis indicated that sheepgrass is closely related to barley and wheat.
Conclusions: This research has greatly enriched sheepgrass transcriptome resources. The identified stress-related genes will help us to decipher the genetic basis of the environmental and ecological adaptations of this species and will be used to improve wheat and barley crops through hybridization or genetic transformation. The EST-SSRs reported here will be a valuable resource for future gene-phenotype studies and for the molecular breeding of sheepgrass and other Poaceae species.
Conflict of interest statement
Figures
References
-
- Liu ZP, Chen ZY, Pan J, Li XF, Su M, et al. (2008) Phylogenetic relationships in Leymus (Poaceae: Triticeae) revealed by the nuclear ribosomal internal transcribed spacer and chloroplast trnL-F sequences. Mol Phylogenet Evol 46: 278–289. - PubMed
-
- Jiang J, Friebe B, Gill BS (1994) Recent advances in alien gene transfer in wheat. Euphytica 73: 199–212.
-
- Liu X, Shi J, Zhang XY, Ma YS, Jia JZ (2001) Screening salt tolerance germplasms and tagging the tolerance gene(s) using microsatellite (SSR) markers in wheat. Acta Bot Sinica 43: 948–954.
-
- Chen PD, Liu WX, Yuan JH, Wang XE, Zhou B, et al. (2005) Development and characterization of wheat-Leymus racemosus translocation lines with resistance to Fusarium Head Blight. Theor Appl Genet 111: 941–948. - PubMed
-
- Qi LL, Pumphrey MO, Briebe B, Chen PD, Gill BS (2008) Molecular cytogenetic characterization of alien introgressions with gene Fhb3 for resistance to Fusarium head blight disease of wheat. Theor Appl Genet 117: 1155–1166. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

