A huntingtin peptide inhibits polyQ-huntingtin associated defects
- PMID: 23861941
- PMCID: PMC3701666
- DOI: 10.1371/journal.pone.0068775
A huntingtin peptide inhibits polyQ-huntingtin associated defects
Abstract
Background: Huntington's disease (HD) is caused by the abnormal expansion of the polyglutamine tract in the human Huntingtin protein (polyQ-hHtt). Although this mutation behaves dominantly, huntingtin loss of function also contributes to HD pathogenesis. Indeed, wild-type Huntingtin plays a protective role with respect to polyQ-hHtt induced defects.
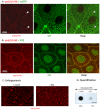
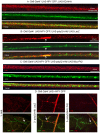
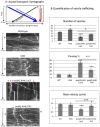
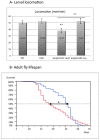
Methodology/principal findings: The question that we addressed here is what part of the wild-type Huntingtin is responsible for these protective properties. We first screened peptides from the Huntingtin protein in HeLa cells and identified a 23 aa peptide (P42) that inhibits polyQ-hHtt aggregation. P42 is part of the endogenous Huntingtin protein and lies within a region rich in proteolytic sites that plays a critical role in the pathogenesis process. Using a Drosophila model of HD, we tested the protective properties of this peptide on aggregation, as well as on different polyQ-hHtt induced neuronal phenotypes: eye degeneration (an indicator of cell death), impairment of vesicular axonal trafficking, and physiological behaviors such as larval locomotion and adult survival. Together, our results demonstrate high protective properties for P42 in vivo, in whole animals. These data also demonstrate a specific role of P42 on Huntington's disease model, since it has no effect on other models of polyQ-induced diseases, such as spinocerebellar ataxias.
Conclusions/significance: Altogether our data show that P42, a 23 aa-long hHtt peptide, plays a protective role with respect to polyQ-hHtt aggregation as well as cellular and behavioral dysfunctions induced by polyQ-hHtt in vivo. Our study also confirms the correlation between polyQ-hHtt aggregation and neuronal defects. Finally, these results strongly suggest a therapeutic potential for P42, specific of Huntington's disease.
Conflict of interest statement
Figures


References
-
- Cowin RM, Roscic A, Bui N, Graham D, Paganetti P, et al. (2012) Neuronal aggregates are associated with phenotypic onset in the R6/2 Huntington's disease transgenic mouse. Behav Brain Res 229: 308–319. - PubMed
-
- Yamamoto A, Lucas JJ, Hen R (2000) Reversal of neuropathology and motor dysfunction in a conditional model of Huntington's disease. Cell 101: 57–66. - PubMed
-
- Goldberg YP, Nicholson DW, Rasper DM, Kalchman MA, Koide HB, et al. (1996) Cleavage of huntingtin by apopain, a proapoptotic cysteine protease, is modulated by the polyglutamine tract. Nat Genet 13: 442–449. - PubMed
-
- Martindale D, Hackam A, Wieczorek A, Ellerby L, Wellington C, et al. (1998) Length of huntingtin and its polyglutamine tract influences localization and frequency of intracellular aggregates. Nat Genet 18: 150–154. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

