The C-terminus of S. pombe DDK subunit Dfp1 is required for meiosis-specific transcription and cohesin cleavage
- PMID: 23862021
- PMCID: PMC3711041
- DOI: 10.1242/bio.20135173
The C-terminus of S. pombe DDK subunit Dfp1 is required for meiosis-specific transcription and cohesin cleavage
Abstract
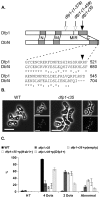
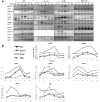
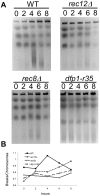
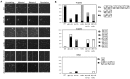
The DDK complex is a conserved kinase complex, consisting of a catalytic subunit, Hsk1 (Cdc7), and its regulatory subunit Dfp1 (Dbf4). This kinase is essential for DNA replication. In this work, we show that dfp1-r35, which truncates the Dfp1 C-terminus zinc finger, causes severe meiotic defects, including reduced spore viability, reduced formation of programmed double strand breaks, altered expression of meiotic genes, and disrupted chromosome segregation. There is a high frequency of dyad formation. Mutants are also defective in the phosphorylation and degradation of the meiotic cohesion, Rec8, resulting in a failure to proceed through the MII division. These defects are more pronounced in a haploid meiosis model than in a normal diploid meiosis. Thus, several critical meiotic functions are linked specifically to the C-terminus of Dfp1, which may target specific substrates for phosphorylation by Hsk1.
Keywords: Cdc7; Dbf4; Fission yeast; Hsk1; Meiosis; Rec8.
Conflict of interest statement
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

