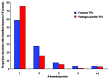
Common and pathogen-specific virulence factors are different in function and structure
- PMID: 23863604
- PMCID: PMC5359729
- DOI: 10.4161/viru.25730
Common and pathogen-specific virulence factors are different in function and structure
Abstract
In the process of host-pathogen interactions, bacterial pathogens always employ some special genes, e.g., virulence factors (VFs) to interact with host and cause damage or diseases to host. A number of VFs have been identified in bacterial pathogens that confer upon bacterial pathogens the ability to cause various types of damage or diseases. However, it has been clarified that some of the identified VFs are also encoded in the genomes of nonpathogenic bacteria, and this finding gives rise to considerable controversy about the definition of virulence factor. Here 1988 virulence factors of 51 sequenced pathogenic bacterial genomes from the virulence factor database (VFDB) were collected, and an orthologous comparison to a non-pathogenic bacteria protein database was conducted using the reciprocal-best-BLAST-hits approach. Six hundred and twenty pathogen-specific VFs and 1368 common VFs (present in both pathogens and nonpathogens) were identified, which account for 31.19% and 68.81% of the total VFs, respectively. The distribution of pathogen-specific VFs and common VFs in pathogenicity islands (PAIs) was systematically investigated, and pathogen-specific VFs were more likely to be located in PAIs than common VFs. The function of the two classes of VFs were also analyzed and compared in depth. Our results indicated that most but not all T3SS proteins are pathogen-specific. T3SS effector proteins tended to be distributed in pathogen-specific VFs, whereas T3SS translocation proteins, apparatus proteins, and chaperones were inclined to be distributed in common VFs. We also observed that exotoxins were located in both pathogen-specific and common VFs. In addition, the architecture of the two classes of VFs was compared, and the results indicated that common VFs had a higher domain number and lower domain coverage value, revealed that common VFs tend to be more complex and less compact proteins.
Keywords: bacterial pathogens; common virulence factor; pathogen-specific virulence factor.
Figures
Comment in
-
Toward a more systematic understanding of bacterial virulence factors and establishing Koch postulates in silico.Virulence. 2013 Aug 15;4(6):437-8. doi: 10.4161/viru.26211. Virulence. 2013. PMID: 23979028 Free PMC article. No abstract available.
References
-
- Binder S, Levitt AM, Sacks JJ, Hughes JM. . Emerging infectious diseases: public health issues for the 21st century. Science 1999; 284:1311 - 3; http://dx.doi.org/ 10.1126/science.284.5418.1311; PMID: 10334978 - DOI - PubMed
-
- Daszak P, Cunningham AA, Hyatt AD. . Emerging infectious diseases of wildlife--threats to biodiversity and human health. Science 2000; 287:443 - 9; http://dx.doi.org/ 10.1126/science.287.5452.443; PMID: 10642539 - DOI - PubMed
-
- Wilson JW, Schurr MJ, LeBlanc CL, Ramamurthy R, Buchanan KL, Nickerson CA. . Mechanisms of bacterial pathogenicity. Postgrad Med J 2002; 78:216 - 24; http://dx.doi.org/ 10.1136/pmj.78.918.216; PMID: 11930024 - DOI - PMC - PubMed
-
- Falkow S. . Molecular Koch’s postulates applied to bacterial pathogenicity--a personal recollection 15 years later. Nat Rev Microbiol 2004; 2:67 - 72; http://dx.doi.org/ 10.1038/nrmicro799; PMID: 15035010 - DOI - PubMed
-
- Wu HJ, Wang AH, Jennings MP. . Discovery of virulence factors of pathogenic bacteria. Curr Opin Chem Biol 2008; 12:93 - 101; http://dx.doi.org/ 10.1016/j.cbpa.2008.01.023; PMID: 18284925 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials