miRISC recruits decapping factors to miRNA targets to enhance their degradation
- PMID: 23863838
- PMCID: PMC3794582
- DOI: 10.1093/nar/gkt619
miRISC recruits decapping factors to miRNA targets to enhance their degradation
Abstract
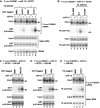
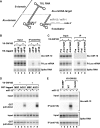
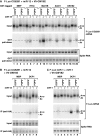
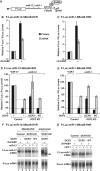
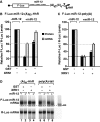
MicroRNA (miRNA)-induced silencing complexes (miRISCs) repress translation and promote degradation of miRNA targets. Target degradation occurs through the 5'-to-3' messenger RNA (mRNA) decay pathway, wherein, after shortening of the mRNA poly(A) tail, the removal of the 5' cap structure by decapping triggers irreversible decay of the mRNA body. Here, we demonstrate that miRISC enhances the association of the decapping activators DCP1, Me31B and HPat with deadenylated miRNA targets that accumulate when decapping is blocked. DCP1 and Me31B recruitment by miRISC occurs before the completion of deadenylation. Remarkably, miRISC recruits DCP1, Me31B and HPat to engineered miRNA targets transcribed by RNA polymerase III, which lack a cap structure, a protein-coding region and a poly(A) tail. Furthermore, miRISC can trigger decapping and the subsequent degradation of mRNA targets independently of ongoing deadenylation. Thus, miRISC increases the local concentration of the decapping machinery on miRNA targets to facilitate decapping and irreversibly shut down their translation.
Figures

References
-
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11:597–610. - PubMed
-
- Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011;12:99–110. - PubMed
-
- Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat. Struct. Mol. Biol. 2012;19:586–593. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

