CELDA -- an ontology for the comprehensive representation of cells in complex systems
- PMID: 23865855
- PMCID: PMC3722091
- DOI: 10.1186/1471-2105-14-228
CELDA -- an ontology for the comprehensive representation of cells in complex systems
Abstract
Background: The need for detailed description and modeling of cells drives the continuous generation of large and diverse datasets. Unfortunately, there exists no systematic and comprehensive way to organize these datasets and their information. CELDA (Cell: Expression, Localization, Development, Anatomy) is a novel ontology for the association of primary experimental data and derived knowledge to various types of cells of organisms.
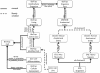
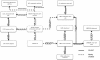
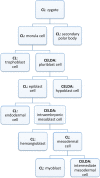
Results: CELDA is a structure that can help to categorize cell types based on species, anatomical localization, subcellular structures, developmental stages and origin. It targets cells in vitro as well as in vivo. Instead of developing a novel ontology from scratch, we carefully designed CELDA in such a way that existing ontologies were integrated as much as possible, and only minimal extensions were performed to cover those classes and areas not present in any existing model. Currently, ten existing ontologies and models are linked to CELDA through the top-level ontology BioTop. Together with 15.439 newly created classes, CELDA contains more than 196.000 classes and 233.670 relationship axioms. CELDA is primarily used as a representational framework for modeling, analyzing and comparing cells within and across species in CellFinder, a web based data repository on cells (http://cellfinder.org).
Conclusions: CELDA can semantically link diverse types of information about cell types. It has been integrated within the research platform CellFinder, where it exemplarily relates cell types from liver and kidney during development on the one hand and anatomical locations in humans on the other, integrating information on all spatial and temporal stages. CELDA is available from the CellFinder website: http://cellfinder.org/about/ontology.
Figures




References
-
- Virchow R. Die Cellularpathologie in ihrer Begründung auf physiologische und pathologische Gewebelehre. Berlin: Verlag von August Hirschwald; 1858.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

