Otx2 and Onecut1 promote the fates of cone photoreceptors and horizontal cells and repress rod photoreceptors
- PMID: 23867227
- PMCID: PMC3819454
- DOI: 10.1016/j.devcel.2013.06.005
Otx2 and Onecut1 promote the fates of cone photoreceptors and horizontal cells and repress rod photoreceptors
Abstract
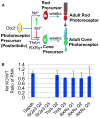
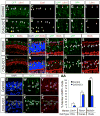
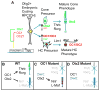
Cone photoreceptors carry out phototransduction in daylight conditions and provide the critical first step in color vision. Despite their importance, little is known about the developmental mechanisms involved in their generation, particularly how they are determined relative to rod photoreceptors, the cells that initiate vision in dim light. Here, we report the identification of a cis-regulatory module (CRM) for the thyroid hormone receptor beta (Thrb) gene, an early cone marker. We found that ThrbCRM1 is active in progenitor cells biased to the production of cones and an interneuronal cell type, the horizontal cell (HC). Molecular analysis of ThrbCRM1 revealed that it is combinatorially regulated by the Otx2 and Onecut1 transcription factors. Onecut1 is sufficient to induce cells with the earliest markers of cones and HCs. Conversely, interference with Onecut1 transcriptional activity leads to precocious rod development, suggesting that Onecut1 is critically important in defining cone versus rod fates.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures




References
-
- Albritton LM, Tseng L, Scadden D, Cunningham JM. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell. 1989;57:659–666. - PubMed
-
- Carter-Dawson LD, LaVail MM. Rods and cones in the mouse retina. II Autoradiographic analysis of cell generation using tritiated thymidine. J Comp Neurol. 1979;188:263–272. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

