Aminoacylating urzymes challenge the RNA world hypothesis
- PMID: 23867455
- PMCID: PMC3772232
- DOI: 10.1074/jbc.M113.496125
Aminoacylating urzymes challenge the RNA world hypothesis
Abstract
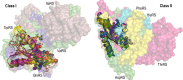
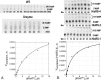
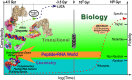
We describe experimental evidence that ancestral peptide catalysts substantially accelerated development of genetic coding. Structurally invariant 120-130-residue Urzymes (Ur = primitive plus enzyme) derived from Class I and Class II aminoacyl-tRNA synthetases (aaRSs) acylate tRNA far faster than the uncatalyzed rate of nonribosomal peptide bond formation from activated amino acids. These new data allow us to demonstrate statistically indistinguishable catalytic profiles for Class I and II aaRSs in both amino acid activation and tRNA acylation, over a time period extending to well before the assembly of full-length enzymes and even further before the Last Universal Common Ancestor. Both Urzymes also exhibit ∼60% of the contemporary catalytic proficiencies. Moreover, they are linked by ancestral sense/antisense genetic coding, and their evident modularities suggest descent from even simpler ancestral pairs also coded by opposite strands of the same gene. Thus, aaRS Urzymes substantially pre-date modern aaRS but are, nevertheless, highly evolved. Their unexpectedly advanced catalytic repertoires, sense/antisense coding, and ancestral modularities imply considerable prior protein-tRNA co-evolution. Further, unlike ribozymes that motivated the RNA World hypothesis, Class I and II Urzyme·tRNA pairs represent consensus ancestral forms sufficient for codon-directed synthesis of nonrandom peptides. By tracing aaRS catalytic activities back to simpler ancestral peptides, we demonstrate key steps for a simpler and hence more probable peptide·RNA development of rapid coding systems matching amino acids with anticodon trinucleotides.
Keywords: Aminoacyl tRNA Synthesis; Aminoacyl tRNA Synthetase; Catalytic RNA; Enzyme Catalysis; Enzyme Proficiency; Molecular Evolution; Origin of Translation; Protein Synthesis; tRNA.
Figures



References
-
- Gilbert W. (1986) Nature 319, 618
-
- Yarus M. (2011) Life from an RNA World: The Ancestor Within, pp. 132–155, Harvard University Press, Cambridge, MA
-
- Koonin E. V. (2012) The Logic of Chance: The Nature and Origin of Biological Evolution, pp. 351–395, Pearson Education, FT Press Science, Upper Saddle River, NJ
-
- Augustine J., Francklyn C. (1997) Design of an active fragment of a class II aminoacyl-tRNA synthetase and its significance for synthetase evolution. Biochemistry 36, 3473–3482 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

