Acinetobacter phage genome is similar to Sphinx 2.36, the circular DNA copurified with TSE infected particles
- PMID: 23867905
- PMCID: PMC3715749
- DOI: 10.1038/srep02240
Acinetobacter phage genome is similar to Sphinx 2.36, the circular DNA copurified with TSE infected particles
Abstract
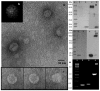
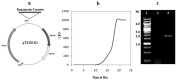
While analyzing plasmids of Acinetobacter sp. DS002 we have detected a circular DNA molecule pTS236, which upon further investigation is identified as the genome of a phage. The phage genome has shown sequence similarity to the recently discovered Sphinx 2.36 DNA sequence co-purified with the Transmissible Spongiform Encephalopathy (TSE) particles isolated from infected brain samples collected from diverse geographical regions. As in Sphinx 2.36, the phage genome also codes for three proteins. One of them codes for RepA and is shown to be involved in replication of pTS236 through rolling circle (RC) mode. The other two translationally coupled ORFs, orf106 and orf96, code for coat proteins of the phage. Although an orf96 homologue was not previously reported in Sphinx 2.36, a closer examination of DNA sequence of Sphinx 2.36 revealed its presence downstream of orf106 homologue. TEM images and infection assays revealed existence of phage AbDs1 in Acinetobacter sp. DS002.
Figures





References
-
- Reams A. B. & Neidle E. L. Genome plasticity in Acinetobacter: new degradative capabilities acquired by the spontaneous amplification of large chromosomal segments. Mol Microbiol 47, 1291–1304 (2003). - PubMed
-
- Fischer R., Bleichrodt F. S. & Gerischer U. C. Aromatic degradative pathways in Acinetobacter baylyi underlie carbon catabolite repression. Microbiology 154, 3095–3103 (2008). - PubMed
-
- Towner K. J. Acinetobacter: an old friend, but a new enemy. J Hosp Infect 73, 355–363 (2009). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

