Glucose starvation induces cell death in K-ras-transformed cells by interfering with the hexosamine biosynthesis pathway and activating the unfolded protein response
- PMID: 23868065
- PMCID: PMC3730427
- DOI: 10.1038/cddis.2013.257
Glucose starvation induces cell death in K-ras-transformed cells by interfering with the hexosamine biosynthesis pathway and activating the unfolded protein response
Erratum in
- Cell Death Dis. 2013;4:e910. Cammarata, F [corrected to Cammarata, F P]
Abstract
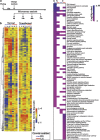
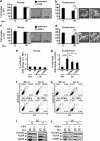
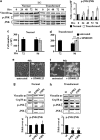
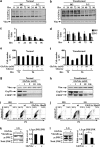
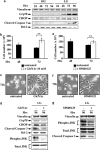
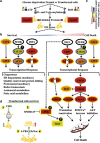
Cancer cells, which use more glucose than normal cells and accumulate extracellular lactate even under normoxic conditions (Warburg effect), have been reported to undergo cell death under glucose deprivation, whereas normal cells remain viable. As it may be relevant to exploit the molecular mechanisms underlying this biological response to achieve new cancer therapies, in this paper we sought to identify them by using transcriptome and proteome analysis applied to an established glucose-addicted cellular model of transformation, namely, murine NIH-3T3 fibroblasts harboring an oncogenic K-RAS gene, compared with parental cells. Noteworthy is that the analyses performed in high- and low-glucose cultures indicate that reduction of glucose availability induces, especially in transformed cells, a significant increase in the expression of several unfolded protein response (UPR) hallmark genes. We show that this response is strictly associated with transformed cell death, given that its attenuation, by reducing protein translation or by increasing cell protein folding capacity, preserves the survival of transformed cells. Such an effect is also observed by inhibiting c-Jun NH2-terminal kinase, a pro-apoptotic signaling mediator set downstream of UPR. Strikingly, addition of N-acetyl-D-glucosamine, a specific substrate for the hexosamine biosynthesis pathway (HBP), to glucose-depleted cells completely prevents transformed cell death, stressing the important role of glucose in HBP fuelling to ensure UPR attenuation and increased cell survival. Interestingly, these results have been fully recognized in a human model of breast cancer, MDA-MB-231 cells. In conclusion, we show that glucose deprivation, leading to harmful accumulation of unfolded proteins in consequence of a reduction of protein glycosylation, induces a UPR-dependent cell death mechanism. These findings may open the way for new therapeutic strategies to specifically kill glycolytic cancer cells.
Figures


References
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell Mar. 2011;144:646–674. - PubMed
-
- Chiaradonna F, Moresco RM, Airoldi C, Gaglio D, Palorini R, Nicotra F, et al. From cancer metabolism to new biomarkers and drug targets. Biotechnol Adv. 2011;30:30–51. - PubMed
-
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. - PubMed
-
- Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–707. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

