Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta
- PMID: 23869020
- PMCID: PMC3736355
- DOI: 10.1126/science.1235872
Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta
Abstract
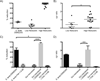
Despite numerous examples of the effects of the human gastrointestinal microbiome on drug efficacy and toxicity, there is often an incomplete understanding of the underlying mechanisms. Here, we dissect the inactivation of the cardiac drug digoxin by the gut Actinobacterium Eggerthella lenta. Transcriptional profiling, comparative genomics, and culture-based assays revealed a cytochrome-encoding operon up-regulated by digoxin, inhibited by arginine, absent in nonmetabolizing E. lenta strains, and predictive of digoxin inactivation by the human gut microbiome. Pharmacokinetic studies using gnotobiotic mice revealed that dietary protein reduces the in vivo microbial metabolism of digoxin, with significant changes to drug concentration in the serum and urine. These results emphasize the importance of viewing pharmacology from the perspective of both our human and microbial genomes.
Figures


References
-
- Haiser HJ, Turnbaugh PJ. Is it time for a metagenomic basis of therapeutics? Science. 2012;336:1253–1255. - PubMed
-
- Goodman LS, et al. Goodman & Gilman's the pharmacological basis of therapeutics. ed. 12th. New York: McGraw-Hill; 2011.
-
- Lindenbaum J, et al. Inactivation of digoxin by the gut flora: reversal by antibiotic therapy. N. Engl. J. Med. 1981;305:789–794. - PubMed
-
- Farr CD, et al. Three-dimensional quantitative structure-activity relationship study of the inhibition of Na(+),K(+)-ATPase by cardiotonic steroids using comparative molecular field analysis. Biochemistry. 2002;41:1137–1148. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

