Time course of gene expression during mouse skeletal muscle hypertrophy
- PMID: 23869057
- PMCID: PMC3798821
- DOI: 10.1152/japplphysiol.00611.2013
Time course of gene expression during mouse skeletal muscle hypertrophy
Abstract
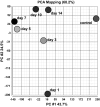
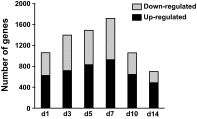
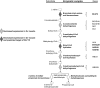
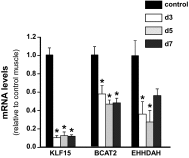
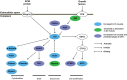
The purpose of this study was to perform a comprehensive transcriptome analysis during skeletal muscle hypertrophy to identify signaling pathways that are operative throughout the hypertrophic response. Global gene expression patterns were determined from microarray results on days 1, 3, 5, 7, 10, and 14 during plantaris muscle hypertrophy induced by synergist ablation in adult mice. Principal component analysis and the number of differentially expressed genes (cutoffs ≥2-fold increase or ≥50% decrease compared with control muscle) revealed three gene expression patterns during overload-induced hypertrophy: early (1 day), intermediate (3, 5, and 7 days), and late (10 and 14 days) patterns. Based on the robust changes in total RNA content and in the number of differentially expressed genes, we focused our attention on the intermediate gene expression pattern. Ingenuity Pathway Analysis revealed a downregulation of genes encoding components of the branched-chain amino acid degradation pathway during hypertrophy. Among these genes, five were predicted by Ingenuity Pathway Analysis or previously shown to be regulated by the transcription factor Kruppel-like factor-15, which was also downregulated during hypertrophy. Moreover, the integrin-linked kinase signaling pathway was activated during hypertrophy, and the downregulation of muscle-specific micro-RNA-1 correlated with the upregulation of five predicted targets associated with the integrin-linked kinase pathway. In conclusion, we identified two novel pathways that may be involved in muscle hypertrophy, as well as two upstream regulators (Kruppel-like factor-15 and micro-RNA-1) that provide targets for future studies investigating the importance of these pathways in muscle hypertrophy.
Keywords: ILK signaling; KLF15; branched-chain amino acid; micro-RNA-1; transcriptome.
Figures


References
-
- Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ, Yancopoulos GD. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol 3: 1014–1019, 2001 - PubMed
-
- Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, Chen RO, Brownstein BH, Cobb JP, Tschoeke SK, Miller-Graziano C, Moldawer LL, Mindrinos MN, Davis RW, Tompkins RG, Lowry SF. A network-based analysis of systemic inflammation in humans. Nature 437: 1032–1037, 2005 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

