Mining differential top-k co-expression patterns from time course comparative gene expression datasets
- PMID: 23870110
- PMCID: PMC3751367
- DOI: 10.1186/1471-2105-14-230
Mining differential top-k co-expression patterns from time course comparative gene expression datasets
Abstract
Background: Frequent pattern mining analysis applied on microarray dataset appears to be a promising strategy for identifying relationships between gene expression levels. Unfortunately, too many itemsets (co-expressed genes) are identified by this analysis method since it does not consider the importance of each gene within biological processes to a cellular response and does not take into account temporal properties under biological treatment-control matched conditions in a microarray dataset.
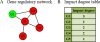
Results: We propose a method termed TIIM (Top-k Impactful Itemsets Miner), which only requires specifying a user-defined number k to explore the top k itemsets with the most significantly differentially co-expressed genes between 2 conditions in a time course. To give genes different weights, a table with impact degrees for each gene was constructed based on the number of neighboring genes that are differently expressed in the dataset within gene regulatory networks. Finally, the resulting top-k impactful itemsets were manually evaluated using previous literature and analyzed by a Gene Ontology enrichment method.
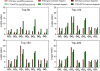
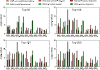
Conclusions: In this study, the proposed method was evaluated in 2 publicly available time course microarray datasets with 2 different experimental conditions. Both datasets identified potential itemsets with co-expressed genes evaluated from the literature and showed higher accuracies compared to the 2 corresponding control methods: i) performing TIIM without considering the gene expression differentiation between 2 different experimental conditions and impact degrees, and ii) performing TIIM with a constant impact degree for each gene. Our proposed method found that several new gene regulations involved in these itemsets were useful for biologists and provided further insights into the mechanisms underpinning biological processes. The Java source code and other related materials used in this study are available at "http://websystem.csie.ncku.edu.tw/TIIM_Program.rar".
Figures




Similar articles
-
An efficient method for mining cross-timepoint gene regulation sequential patterns from time course gene expression datasets.BMC Bioinformatics. 2013;14 Suppl 12(Suppl 12):S3. doi: 10.1186/1471-2105-14-S12-S3. Epub 2013 Sep 24. BMC Bioinformatics. 2013. PMID: 24267918 Free PMC article.
-
Mining significant high utility gene regulation sequential patterns.BMC Syst Biol. 2017 Dec 14;11(Suppl 6):109. doi: 10.1186/s12918-017-0475-4. BMC Syst Biol. 2017. PMID: 29297335 Free PMC article.
-
Differential regulation enrichment analysis via the integration of transcriptional regulatory network and gene expression data.Bioinformatics. 2015 Feb 15;31(4):563-71. doi: 10.1093/bioinformatics/btu672. Epub 2014 Oct 15. Bioinformatics. 2015. PMID: 25322838
-
Efficient Top-K Identical Frequent Itemsets Mining without Support Threshold Parameter from Transactional Datasets Produced by IoT-Based Smart Shopping Carts.Sensors (Basel). 2022 Oct 21;22(20):8063. doi: 10.3390/s22208063. Sensors (Basel). 2022. PMID: 36298424 Free PMC article.
-
Multiobjective triclustering of time-series transcriptome data reveals key genes of biological processes.BMC Bioinformatics. 2015 Jun 26;16:200. doi: 10.1186/s12859-015-0635-8. BMC Bioinformatics. 2015. PMID: 26108437 Free PMC article.
Cited by
-
A multi-objective gene clustering algorithm guided by apriori biological knowledge with intensification and diversification strategies.BioData Min. 2018 Aug 7;11:16. doi: 10.1186/s13040-018-0178-4. eCollection 2018. BioData Min. 2018. PMID: 30100924 Free PMC article.
-
A hybrid multi-objective whale optimization algorithm for analyzing microarray data based on Apache Spark.PeerJ Comput Sci. 2021 Mar 25;7:e416. doi: 10.7717/peerj-cs.416. eCollection 2021. PeerJ Comput Sci. 2021. PMID: 33834101 Free PMC article.
-
Practical Approaches for Mining Frequent Patterns in Molecular Datasets.Bioinform Biol Insights. 2016 May 2;10:37-47. doi: 10.4137/BBI.S38419. eCollection 2016. Bioinform Biol Insights. 2016. PMID: 27168722 Free PMC article. Review.
-
eXplainable Artificial Intelligence (XAI) for the identification of biologically relevant gene expression patterns in longitudinal human studies, insights from obesity research.PLoS Comput Biol. 2020 Apr 10;16(4):e1007792. doi: 10.1371/journal.pcbi.1007792. eCollection 2020 Apr. PLoS Comput Biol. 2020. PMID: 32275707 Free PMC article.
-
MiningABs: mining associated biomarkers across multi-connected gene expression datasets.BMC Bioinformatics. 2014 Jun 8;15:173. doi: 10.1186/1471-2105-15-173. BMC Bioinformatics. 2014. PMID: 24909518 Free PMC article.
References
-
- McIntosh T, Chawla S. High confidence rule mining for microarray analysis. IEEE/ACM transactions on computational biology and bioinformatics / IEEE, ACM. 2007;4(4):611–623. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

