An epigenetic trap stabilizes singular olfactory receptor expression
- PMID: 23870122
- PMCID: PMC3929589
- DOI: 10.1016/j.cell.2013.06.039
An epigenetic trap stabilizes singular olfactory receptor expression
Abstract
The molecular mechanisms regulating olfactory receptor (OR) expression in the mammalian nose are not yet understood. Here, we identify the transient expression of histone demethylase LSD1 and the OR-dependent expression of adenylyl cyclase 3 (Adcy3) as requirements for initiation and stabilization of OR expression. As a transcriptional coactivator, LSD1 is necessary for desilencing and initiating OR transcription, but as a transcriptional corepressor, it is incompatible with maintenance of OR expression, and its downregulation is imperative for stable OR choice. Adcy3, a sensor of OR expression and a transmitter of an OR-elicited feedback, mediates the downregulation of LSD1 and promotes the differentiation of olfactory sensory neurons (OSNs). This novel, three-node signaling cascade locks the epigenetic state of the chosen OR, stabilizes its singular expression, and prevents the transcriptional activation of additional OR alleles for the life of the neuron.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
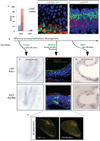
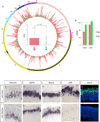
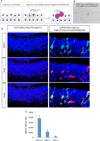

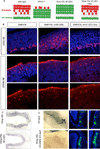

Comment in
-
An epigenetic trap involved in olfactory receptor gene choice.Dev Cell. 2013 Jul 29;26(2):120-1. doi: 10.1016/j.devcel.2013.07.011. Dev Cell. 2013. PMID: 23906063
-
Sensory systems: Making a choice and sticking with it.Nat Rev Neurosci. 2013 Sep;14(9):590-1. doi: 10.1038/nrn3572. Epub 2013 Aug 7. Nat Rev Neurosci. 2013. PMID: 23921412 No abstract available.
References
-
- Alon U. Network motifs: theory and experimental approaches. Nature reviews. 2007;8:450–461. - PubMed
-
- Anand R, Marmorstein R. Structure and mechanism of lysine-specific demethylase enzymes. The Journal of biological chemistry. 2007;282:35425–35429. - PubMed
-
- Barnea G, O'Donnell S, Mancia F, Sun X, Nemes A, Mendelsohn M, Axel R. Odorant receptors on axon termini in the brain. Science (New York, NY. 2004;304:1468. - PubMed
-
- Belluscio L, Gold GH, Nemes A, Axel R. Mice deficient in G(olf) are anosmic. Neuron. 1998;20:69–81. - PubMed
-
- Brunet LJ, Gold GH, Ngai J. General anosmia caused by a targeted disruption of the mouse olfactory cyclic nucleotide-gated cation channel. Neuron. 1996;17:681–693. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

