Cortical dynein and asymmetric membrane elongation coordinately position the spindle in anaphase
- PMID: 23870127
- PMCID: PMC4177044
- DOI: 10.1016/j.cell.2013.06.010
Cortical dynein and asymmetric membrane elongation coordinately position the spindle in anaphase
Erratum in
- Cell. 2013 Sep 12;154(6):1401
Abstract
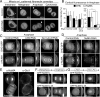
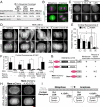
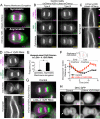
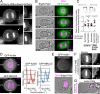
Mitotic spindle position defines the cell-cleavage site during cytokinesis. However, the mechanisms that control spindle positioning to generate equal-sized daughter cells remain poorly understood. Here, we demonstrate that two mechanisms act coordinately to center the spindle during anaphase in symmetrically dividing human cells. First, the spindle is positioned directly by the microtubule-based motor dynein, which we demonstrate is targeted to the cell cortex by two distinct pathways: a Gαi/LGN/NuMA-dependent pathway and a 4.1G/R and NuMA-dependent, anaphase-specific pathway. Second, we find that asymmetric plasma membrane elongation occurs in response to spindle mispositioning to alter the cellular boundaries relative to the spindle. Asymmetric membrane elongation is promoted by chromosome-derived Ran-GTP signals that locally reduce Anillin at the growing cell cortex. In asymmetrically elongating cells, dynein-dependent spindle anchoring at the stationary cell cortex ensures proper spindle positioning. Our results reveal the anaphase-specific spindle centering systems that achieve equal-sized cell division.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures


Comment in
-
Born equal: dual safeguards for daughter cell size symmetry.Cell. 2013 Jul 18;154(2):269-71. doi: 10.1016/j.cell.2013.06.035. Cell. 2013. PMID: 23870117
-
Cell cycle: Getting to the centre.Nat Rev Mol Cell Biol. 2013 Sep;14(9):545. doi: 10.1038/nrm3648. Epub 2013 Aug 14. Nat Rev Mol Cell Biol. 2013. PMID: 23942452 No abstract available.
References
-
- Bennett V, Baines AJ. Spectrin and ankyrin-based pathways: metazoan inventions for integrating cells into tissues. Physiological reviews. 2001;81:1353–1392. - PubMed
-
- Burgess DR, Chang F. Site selection for the cleavage furrow at cytokinesis. Trends Cell Biol. 2005;15:156–162. - PubMed
-
- Charras G, Paluch E. Blebs lead the way: how to migrate without lamellipodia. Nat Rev Mol Cell Biol. 2008;9:730–736. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

