A microscale human liver platform that supports the hepatic stages of Plasmodium falciparum and vivax
- PMID: 23870318
- PMCID: PMC3780791
- DOI: 10.1016/j.chom.2013.06.005
A microscale human liver platform that supports the hepatic stages of Plasmodium falciparum and vivax
Abstract
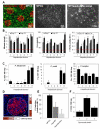
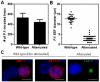
The Plasmodium liver stage is an attractive target for the development of antimalarial drugs and vaccines, as it provides an opportunity to interrupt the life cycle of the parasite at a critical early stage. However, targeting the liver stage has been difficult. Undoubtedly, a major barrier has been the lack of robust, reliable, and reproducible in vitro liver-stage cultures. Here, we establish the liver stages for both Plasmodium falciparum and Plasmodium vivax in a microscale human liver platform composed of cryopreserved, micropatterned human primary hepatocytes surrounded by supportive stromal cells. Using this system, we have successfully recapitulated the full liver stage of P. falciparum, including the release of infected merozoites and infection of overlaid erythrocytes, as well as the establishment of small forms in late liver stages of P. vivax. Finally, we validate the potential of this platform as a tool for medium-throughput antimalarial drug screening and vaccine development.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures




Comment in
-
Techniques & applications: A new tool for liver-stage malaria.Nat Rev Microbiol. 2013 Sep;11(9):596. doi: 10.1038/nrmicro3097. Epub 2013 Aug 5. Nat Rev Microbiol. 2013. PMID: 23912212 No abstract available.
References
-
- Annoura T, Ploemen IH, van Schaijk BC, Sajid M, Vos MW, van Gemert GJ, Chevalley-Maurel S, Franke-Fayard BM, Hermsen CC, Gego A, et al. Assessing the adequacy of attenuation of genetically modified malaria parasite vaccine candidates. Vaccine. 2012;30:2662–2670. - PubMed
-
- Bhatia SN, Balis UJ, Yarmush ML, Toner M. Effect of cell-cell interactions in preservation of cellular phenotype: cocultivation of hepatocytes and nonparenchymal cells. FASEB J. 1999;13:1883–1900. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

