Imprinted chromatin around DIRAS3 regulates alternative splicing of GNG12-AS1, a long noncoding RNA
- PMID: 23871723
- PMCID: PMC3738830
- DOI: 10.1016/j.ajhg.2013.06.010
Imprinted chromatin around DIRAS3 regulates alternative splicing of GNG12-AS1, a long noncoding RNA
Abstract
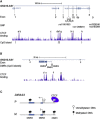
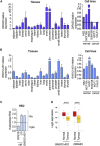
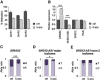
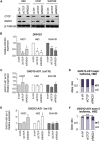
Imprinted gene clusters are regulated by long noncoding RNAs (lncRNAs), CCCTC binding factor (CTCF)-mediated boundaries, and DNA methylation. DIRAS3 (also known as ARH1 or NOEY1) is an imprinted gene encoding a protein belonging to the RAS superfamily of GTPases and is located within an intron of a lncRNA called GNG12-AS1. In this study, we investigated whether GNG12-AS1 is imprinted and coregulated with DIRAS3. We report that GNG12-AS1 is coexpressed with DIRAS3 in several tissues and coordinately downregulated with DIRAS3 in breast cancers. GNG12-AS1 has several splice variants, all of which initiate from a single transcription start site. In placenta tissue and normal cell lines, GNG12-AS1 is biallelically expressed but some isoforms are allele-specifically spliced. Cohesin plays a role in allele-specific splicing of GNG12-AS1. In breast cancer cell lines with loss of DIRAS3 imprinting, DIRAS3 and GNG12-AS1 are silenced in cis and the remaining GNG12-AS1 transcripts are predominantly monoallelic. The GNG12-AS1 locus, which includes DIRAS3, provides an example of imprinted cotranscriptional splicing and a potential model system for studying the long-range effects of CTCF-cohesin binding on splicing and transcriptional interference.
Copyright © 2013 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Wang L., Hoque A., Luo R.Z., Yuan J., Lu Z., Nishimoto A., Liu J., Sahin A.A., Lippman S.M., Bast R.C., Jr., Yu Y. Loss of the expression of the tumor suppressor gene ARHI is associated with progression of breast cancer. Clin. Cancer Res. 2003;9:3660–3666. - PubMed
-
- Klingauf M., Beck M., Berge U., Turgay Y., Heinzer S., Horvath P., Kroschewski R. The tumour suppressor DiRas3 interacts with C-RAF and downregulates MEK activity to restrict cell migration. Biol. Cell. 2013;105:91–107. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

