A comparison of in vitro nucleosome positioning mapped with chicken, frog and a variety of yeast core histones
- PMID: 23871836
- PMCID: PMC3899014
- DOI: 10.1016/j.jmb.2013.07.019
A comparison of in vitro nucleosome positioning mapped with chicken, frog and a variety of yeast core histones
Abstract
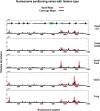
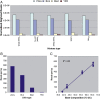
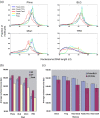
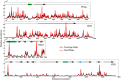
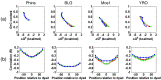
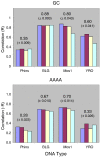
Using high-throughput sequencing, we have mapped sequence-directed nucleosome positioning in vitro on four plasmid DNAs containing DNA fragments derived from the genomes of sheep, drosophila, human and yeast. Chromatins were prepared by reconstitution using chicken, frog and yeast core histones. We also assembled yeast chromatin in which histone H3 was replaced by the centromere-specific histone variant, Cse4. The positions occupied by recombinant frog and native chicken histones were found to be very similar. In contrast, nucleosomes containing the canonical yeast octamer or, in particular, the Cse4 octamer were assembled at distinct populations of locations, a property that was more apparent on particular genomic DNA fragments. The factors that may contribute to this variation in nucleosome positioning and the implications of the behavior are discussed.
Keywords: Cse4; chromatin; core histones; histone variants; nucleosome positioning.
© 2013. Published by Elsevier Ltd. All rights reserved.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

