High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy
- PMID: 23872039
- PMCID: PMC3834153
- DOI: 10.1016/j.ultramic.2013.06.004
High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy
Abstract
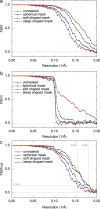
Three-dimensional (3D) structure determination by single particle electron cryomicroscopy (cryoEM) involves the calculation of an initial 3D model, followed by extensive iterative improvement of the orientation determination of the individual particle images and the resulting 3D map. Because there is much more noise than signal at high resolution in the images, this creates the possibility of noise reinforcement in the 3D map, which can give a false impression of the resolution attained. The balance between signal and noise in the final map at its limiting resolution depends on the image processing procedure and is not easily predicted. There is a growing awareness in the cryoEM community of how to avoid such over-fitting and over-estimation of resolution. Equally, there has been a reluctance to use the two principal methods of avoidance because they give lower resolution estimates, which some people believe are too pessimistic. Here we describe a simple test that is compatible with any image processing protocol. The test allows measurement of the amount of signal and the amount of noise from overfitting that is present in the final 3D map. We have applied the method to two different sets of cryoEM images of the enzyme beta-galactosidase using several image processing packages. Our procedure involves substituting the Fourier components of the initial particle image stack beyond a chosen resolution by either the Fourier components from an adjacent area of background, or by simple randomisation of the phases of the particle structure factors. This substituted noise thus has the same spectral power distribution as the original data. Comparison of the Fourier Shell Correlation (FSC) plots from the 3D map obtained using the experimental data with that from the same data with high-resolution noise (HR-noise) substituted allows an unambiguous measurement of the amount of overfitting and an accompanying resolution assessment. A simple formula can be used to calculate an unbiased FSC from the two curves, even when a substantial amount of overfitting is present. The approach is software independent. The user is therefore completely free to use any established method or novel combination of methods, provided the HR-noise test is carried out in parallel. Applying this procedure to cryoEM images of beta-galactosidase shows how overfitting varies greatly depending on the procedure, but in the best case shows no overfitting and a resolution of ~6 Å. (382 words).
Keywords: Beta-galactosidase; Electron cryomicroscopy; Overfitting; Resolution; Single particle; Validation.
© 2013 The Authors. Published by Elsevier B.V. All rights reserved.
Figures




References
-
- Dubochet J., Adrian M., Chang J.J., Homo J.C., Lepault J., McDowall A.W., Schultz P. Cryo-electron microscopy of vitrified specimens. Quarterly Reviews of Biophysics. 1988;21:129–228. - PubMed
-
- Taylor K.A., Glaeser R.M. Electron diffraction of frozen, hydrated protein crystals. Science. 1974;186:1036–1037. - PubMed
-
- Fujiyoshi Y., Mizusaki T., Morikawa K., Yamagishi H., Aoki Y., Kihara H., Harada Y. Development of a superfluid-helium stage for high-resolution electron-microscopy. Ultramicroscopy. 1991;38:241–251.
-
- Zemlin F., Beckmann E., vanderMast K.D. A 200 kV electron microscope with Schottky field emitter and a helium-cooled superconducting objective lens. Ultramicroscopy. 1996;63:227–238.
-
- Homo J.C., Booy F., Labouesse P., Lepault J., Dubochet J. Improved anti-contaminator for cryo-electron microscopy with a Philips EM-400. Journal of Microscopy-Oxford. 1984;136:337–340.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

