Structural insights into the proton pumping by unusual proteorhodopsin from nonmarine bacteria
- PMID: 23872846
- PMCID: PMC3732926
- DOI: 10.1073/pnas.1221629110
Structural insights into the proton pumping by unusual proteorhodopsin from nonmarine bacteria
Erratum in
- Proc Natl Acad Sci U S A. 2013 Sep 3;110(36):14813. Arseniev, Alexander A [corrected to Arseniev, Alexander S]
Abstract
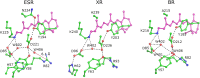
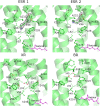
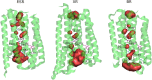
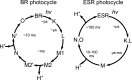
Light-driven proton pumps are present in many organisms. Here, we present a high-resolution structure of a proteorhodopsin from a permafrost bacterium, Exiguobacterium sibiricum rhodopsin (ESR). Contrary to the proton pumps of known structure, ESR possesses three unique features. First, ESR's proton donor is a lysine side chain that is situated very close to the bulk solvent. Second, the α-helical structure in the middle of the helix F is replaced by 3(10)- and π-helix-like elements that are stabilized by the Trp-154 and Asn-224 side chains. This feature is characteristic for the proteorhodopsin family of proteins. Third, the proton release region is connected to the bulk solvent by a chain of water molecules already in the ground state. Despite these peculiarities, the positions of water molecule and amino acid side chains in the immediate Schiff base vicinity are very well conserved. These features make ESR a very unusual proton pump. The presented structure sheds light on the large family of proteorhodopsins, for which structural information was not available previously.
Keywords: bacteriorhodopsin; photocycle; retinal; retinylidene protein.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Spudich JL, Yang C-S, Jung K-H, Spudich EN. Retinylidene proteins: Structures and functions from archaea to humans. Annu Rev Cell Dev Biol. 2000;16:365–392. - PubMed
-
- Pastrana E. Optogenetics: Controlling cell function with light. Nat Methods. 2011;8:24–25.
-
- Oesterhelt D, Stoeckenius W. Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nat New Biol. 1971;233(39):149–152. - PubMed
-
- Béjà O, et al. Bacterial rhodopsin: Evidence for a new type of phototrophy in the sea. Science. 2000;289(5486):1902–1906. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

