Uba1 functions in Atg7- and Atg3-independent autophagy
- PMID: 23873149
- PMCID: PMC3762904
- DOI: 10.1038/ncb2804
Uba1 functions in Atg7- and Atg3-independent autophagy
Abstract
Autophagy is a conserved process that delivers components of the cytoplasm to lysosomes for degradation. The E1 and E2 enzymes encoded by Atg7 and Atg3 are thought to be essential for autophagy involving the ubiquitin-like protein Atg8. Here, we describe an Atg7- and Atg3-independent autophagy pathway that facilitates programmed reduction of cell size during intestine cell death. Although multiple components of the core autophagy pathways, including Atg8, are required for autophagy and cells to shrink in the midgut of the intestine, loss of either Atg7 or Atg3 function does not influence these cellular processes. Rather, Uba1, the E1 enzyme used in ubiquitylation, is required for autophagy and reduction of cell size. Our data reveal that distinct autophagy programs are used by different cells within an animal, and disclose an unappreciated role for ubiquitin activation in autophagy.
Figures


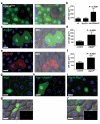
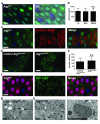
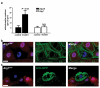
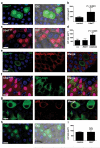

Comment in
-
Autophagy: Atg independence in the midgut.Nat Rev Mol Cell Biol. 2013 Sep;14(9):546. doi: 10.1038/nrm3646. Epub 2013 Aug 7. Nat Rev Mol Cell Biol. 2013. PMID: 23921334 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

