The nuclear envelope LEM-domain protein emerin
- PMID: 23873439
- PMCID: PMC3810338
- DOI: 10.4161/nucl.25751
The nuclear envelope LEM-domain protein emerin
Abstract
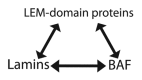
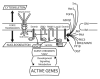
Emerin, a conserved LEM-domain protein, is among the few nuclear membrane proteins for which extensive basic knowledge--biochemistry, partners, functions, localizations, posttranslational regulation, roles in development and links to human disease--is available. This review summarizes emerin and its emerging roles in nuclear "lamina" structure, chromatin tethering, gene regulation, mitosis, nuclear assembly, development, signaling and mechano-transduction. We also highlight many open questions, exploration of which will be critical to understand how this intriguing nuclear membrane protein and its "family" influence the genome.
Keywords: Emery-Dreifuss muscular dystrophy; GAGE cancer-testis antigen; LEM-domain; Nestor-Guillermo progeria; O-GlcNAc transferase; barrier-to-autointegration factor; emerin; laminopathy; nuclear envelope; nucleoskeleton.
Figures



References
-
- Korfali N, Wilkie GS, Swanson SK, Srsen V, Batrakou DG, Fairley EA, et al. The leukocyte nuclear envelope proteome varies with cell activation and contains novel transmembrane proteins that affect genome architecture. Mol Cell Proteomics. 2010;9:2571–85. doi: 10.1074/mcp.M110.002915. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
