DockRank: ranking docked conformations using partner-specific sequence homology-based protein interface prediction
- PMID: 23873600
- PMCID: PMC4417613
- DOI: 10.1002/prot.24370
DockRank: ranking docked conformations using partner-specific sequence homology-based protein interface prediction
Abstract
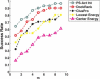
Selecting near-native conformations from the immense number of conformations generated by docking programs remains a major challenge in molecular docking. We introduce DockRank, a novel approach to scoring docked conformations based on the degree to which the interface residues of the docked conformation match a set of predicted interface residues. DockRank uses interface residues predicted by partner-specific sequence homology-based protein-protein interface predictor (PS-HomPPI), which predicts the interface residues of a query protein with a specific interaction partner. We compared the performance of DockRank with several state-of-the-art docking scoring functions using Success Rate (the percentage of cases that have at least one near-native conformation among the top m conformations) and Hit Rate (the percentage of near-native conformations that are included among the top m conformations). In cases where it is possible to obtain partner-specific (PS) interface predictions from PS-HomPPI, DockRank consistently outperforms both (i) ZRank and IRAD, two state-of-the-art energy-based scoring functions (improving Success Rate by up to 4-fold); and (ii) Variants of DockRank that use predicted interface residues obtained from several protein interface predictors that do not take into account the binding partner in making interface predictions (improving success rate by up to 39-fold). The latter result underscores the importance of using partner-specific interface residues in scoring docked conformations. We show that DockRank, when used to re-rank the conformations returned by ClusPro, improves upon the original ClusPro rankings in terms of both Success Rate and Hit Rate. DockRank is available as a server at http://einstein.cs.iastate.edu/DockRank/.
Keywords: docking scoring functions; homo-interologs; partner-specific protein-protein interface residue prediction; protein complex structure prediction; protein-protein docking; sequence homologs.
Copyright © 2013 Wiley Periodicals, Inc.
Figures





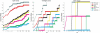
References
-
- Kuzu G, Keskin O, Gursoy A, Nussinov R. Constructing structural networks of signaling pathways on the proteome scale. Curr Opin Struct Biol. 2012;22:367–377. - PubMed
-
- Acuner Ozbabacan S, Engin H, Gursoy A, Keskin O. Transient protein–protein interactions. Protein Eng Des Sel. 2011;24:635–648. - PubMed
-
- Nooren I, Thornton J. Structural characterisation and functional significance of transient protein-protein interactions. J Mol Biol. 2003;325:991–1018. - PubMed
-
- Grosdidier S, Fernandez-Recio J. Protein-protein docking and hot-spot prediction for drug discovery. Curr Pharm Des. 2012;18:4607–4618. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

