Genome-scale co-evolutionary inference identifies functions and clients of bacterial Hsp90
- PMID: 23874229
- PMCID: PMC3708813
- DOI: 10.1371/journal.pgen.1003631
Genome-scale co-evolutionary inference identifies functions and clients of bacterial Hsp90
Abstract
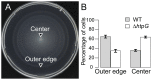
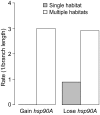
The molecular chaperone Hsp90 is essential in eukaryotes, in which it facilitates the folding of developmental regulators and signal transduction proteins known as Hsp90 clients. In contrast, Hsp90 is not essential in bacteria, and a broad characterization of its molecular and organismal function is lacking. To enable such characterization, we used a genome-scale phylogenetic analysis to identify genes that co-evolve with bacterial Hsp90. We find that genes whose gain and loss were coordinated with Hsp90 throughout bacterial evolution tended to function in flagellar assembly, chemotaxis, and bacterial secretion, suggesting that Hsp90 may aid assembly of protein complexes. To add to the limited set of known bacterial Hsp90 clients, we further developed a statistical method to predict putative clients. We validated our predictions by demonstrating that the flagellar protein FliN and the chemotaxis kinase CheA behaved as Hsp90 clients in Escherichia coli, confirming the predicted role of Hsp90 in chemotaxis and flagellar assembly. Furthermore, normal Hsp90 function is important for wild-type motility and/or chemotaxis in E. coli. This novel function of bacterial Hsp90 agreed with our subsequent finding that Hsp90 is associated with a preference for multiple habitats and may therefore face a complex selection regime. Taken together, our results reveal previously unknown functions of bacterial Hsp90 and open avenues for future experimental exploration by implicating Hsp90 in the assembly of membrane protein complexes and adaptation to novel environments.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Only one of the five CheY homologs in Vibrio cholerae directly switches flagellar rotation.J Bacteriol. 2005 Dec;187(24):8403-10. doi: 10.1128/JB.187.24.8403-8410.2005. J Bacteriol. 2005. PMID: 16321945 Free PMC article.
-
Molecular characterization of a flagellar/chemotaxis operon in the spirochete Borrelia burgdorferi.FEMS Microbiol Lett. 1997 Aug 15;153(2):425-31. doi: 10.1111/j.1574-6968.1997.tb12606.x. FEMS Microbiol Lett. 1997. PMID: 9271872
-
Exploiting genome sequence: predictions for mechanisms of Campylobacter chemotaxis.Trends Microbiol. 2002 Apr;10(4):155-9. doi: 10.1016/s0966-842x(02)02323-5. Trends Microbiol. 2002. PMID: 11912013
-
Evolution and function of diverse Hsp90 homologs and cochaperone proteins.Biochim Biophys Acta. 2012 Mar;1823(3):607-13. doi: 10.1016/j.bbamcr.2011.09.020. Epub 2011 Oct 8. Biochim Biophys Acta. 2012. PMID: 22008467 Review.
-
The role of Hsp90 in protein complex assembly.Biochim Biophys Acta. 2012 Mar;1823(3):674-82. doi: 10.1016/j.bbamcr.2011.09.001. Epub 2011 Sep 16. Biochim Biophys Acta. 2012. PMID: 21945180 Review.
Cited by
-
Pervasive convergent evolution and extreme phenotypes define chaperone requirements of protein homeostasis.Proc Natl Acad Sci U S A. 2019 Oct 1;116(40):20009-20014. doi: 10.1073/pnas.1904611116. Epub 2019 Sep 16. Proc Natl Acad Sci U S A. 2019. PMID: 31527276 Free PMC article.
-
Chance Favors the Prepared Genomes: Horizontal Transfer Shapes the Emergence of Antibiotic Resistance Mutations in Core Genes.Mol Biol Evol. 2023 Oct 4;40(10):msad217. doi: 10.1093/molbev/msad217. Mol Biol Evol. 2023. PMID: 37788575 Free PMC article.
-
Evolutionary assembly patterns of prokaryotic genomes.Genome Res. 2016 Jun;26(6):826-33. doi: 10.1101/gr.200097.115. Epub 2016 Apr 14. Genome Res. 2016. PMID: 27197212 Free PMC article.
-
Physics of swimming and its fitness cost determine strategies of bacterial investment in flagellar motility.Nat Commun. 2025 Feb 18;16(1):1731. doi: 10.1038/s41467-025-56980-x. Nat Commun. 2025. PMID: 39966405 Free PMC article.
-
Bacterial Hsp90 Facilitates the Degradation of Aggregation-Prone Hsp70-Hsp40 Substrates.Front Mol Biosci. 2021 Apr 15;8:653073. doi: 10.3389/fmolb.2021.653073. eCollection 2021. Front Mol Biosci. 2021. PMID: 33937334 Free PMC article.
References
-
- Rutherford SL, Zuker CS (1994) Protein Folding and the Regulation of Signaling Pathways. Cell 79: 1129–1132. - PubMed
-
- Picard D (2002) Heat-shock protein 90, a chaperone for folding and regulation. Cellular and Molecular Life Sciences 59: 1640–1648 doi:10.1007/PL00012491 - DOI - PMC - PubMed
-
- Young JC (2001) Hsp90: a specialized but essential protein-folding tool. The Journal of Cell Biology 154: 267–274 doi:10.1083/jcb.200104079 - DOI - PMC - PubMed
-
- Zhao R, Davey M, Hsu Y-C, Kaplanek P, Tong A, et al. (2005) Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell 120: 715–727 doi:10.1016/j.cell.2004.12.024 - DOI - PubMed
-
- Taipale M, Jarosz DF, Lindquist S (2010) HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nature reviews Molecular cell biology 11: 515–528 doi:10.1038/nrm2918 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

